

林业科学 ›› 2025, Vol. 61 ›› Issue (2): 131-141.doi: 10.11707/j.1001-7488.LYKX20240119
刘明彤,庄和必,王紫彤,石帅宾,刘晓娟,林二培,胡现铬,黄华宏*( )
)
收稿日期:2024-03-01
出版日期:2025-02-25
发布日期:2025-03-03
通讯作者:
黄华宏
E-mail:huanghh1976@163.com
基金资助:
Mingtong Liu,Hebi Zhuang,Zitong Wang,Shuaibin Shi,Xiaojuan Liu,Erpei Lin,Xiange Hu,Huahong Huang*( )
)
Received:2024-03-01
Online:2025-02-25
Published:2025-03-03
Contact:
Huahong Huang
E-mail:huanghh1976@163.com
摘要:
目的: 在挖掘杉木次生壁发育相关NAC调控因子基础上,分析其生物信息学特征、表达模式,以及在拟南芥中过表达的作用,以期为杉木次生壁发育分子机制解析和分子辅助育种提供重要参考价值。方法: 基于杉木不同器官和组织的转录组测序数据,通过共表达网络分析(WGCNA)筛选与木质化相关的NAC基因,克隆其全长序列,进行序列比对和系统进化树分析;利用实时荧光定量PCR技术(RT-qPCR)分析其在不同组织和应压木形成中的表达模式;构建NAC基因过表达重组载体,利用花序侵染法获得转基因拟南芥,并进行转基因植株花序茎横切面染色观察和木质素合成相关基因的表达分析。结果: 分离得到1个杉木NAC基因(ClNAC40),其cDNA序列长度为1 556 bp,开放阅读框(ORF)为1 344 bp,编码447个氨基酸。获得ClNAC40基因组序列共3 430 bp,由6个外显子和5个内含子组成。进化树分析发现ClNAC40与已报道的次生发育相关NAC转录因子聚为一类。ClNAC40基因在木质部(XY)优势表达,而在根(RT)、雌球花(FC)、雄球花(MC)和皮层(BK)中的表达量相对较低;同时,在茎中表达呈现随木质化程度升高而递增趋势,在木质化茎(S3)中的表达量是未木质化茎(S1)的3.2倍。在应压木诱导形成过程中,ClNAC40在应压木中的表达量呈上升趋势,斜放处理30天和60天时的表达水平均显著高于直立木(对照)。在拟南芥中过表达ClNAC40导致花序茎的高度和直径均明显增加;花序茎横切面经间苯三酚染色,显色较野生型深且染色区域占比更大,表明木质素沉积更多。RT-qPCR分析结果表明,ClNAC40过表达显著提高了拟南芥木质素合成途径关键酶基因的表达。结论: 杉木ClNAC40基因通过激活木质素生物合成相关酶基因表达,参与调控次生壁的发育。
中图分类号:
刘明彤,庄和必,王紫彤,石帅宾,刘晓娟,林二培,胡现铬,黄华宏. 杉木次生壁发育调控转录因子基因ClNAC40的生物学功能[J]. 林业科学, 2025, 61(2): 131-141.
Mingtong Liu,Hebi Zhuang,Zitong Wang,Shuaibin Shi,Xiaojuan Liu,Erpei Lin,Xiange Hu,Huahong Huang. Biological Functional Analysis of Transcription Factor Gene ClNAC40 Regulating Secondary Cell Wall Development in Cunninghamia lanceolata[J]. Scientia Silvae Sinicae, 2025, 61(2): 131-141.
表2
荧光定量PCR引物"
| 引物名称 Primer name | 序列 Sequence (5'→3') | 引物名称 Primer name | 序列 Sequence (5'→3') | |
| ClNAC40-QF | TATCAGACAGTGGTAAAGGCT | AtCAD5-QF | TTGGCTGATTCGTTGGATTA | |
| ClNAC40-QR | TATCAGACAGTGGTAAAGGCT | AtCAD5-QR | ATCACTTTCCTCCCAAGCAT | |
| ClActin1-QF | GAGGGACCAGATTCATCGTATTC | AtCOMT1-QF | CTGCTACGAGTCACTTCCAGAGG | |
| ClActin1-QR | ATGCTGGTATTGCTGATCGTATG | AtCOMT1-QR | GAGTAACTCAATAAGGTTAACACC | |
| AtActin2-QF | CCTGAAAGGAAGTACAGTG | AtHCT-QF | GCCTGCACCAAGTATGAAGA | |
| AtActin2-QR | CTGTGAACGATTCCTGGAC | AtHCT-QR | GACAGTGTTCCCATCCTCCT | |
| AtPAL1-QF | AAGATTGGAGCTTTCGAGGA | AtC3H1-QF | GTTGGACTTGACCGGATCTT | |
| AtPAL1-QR | TCTGTTCCAAGCTCTTCCCT | AtC3H1-QR | ATTAGAGGCGTTGGAGGATG | |
| AtPAL2-QF | GAGGCAGCGTTAAGGTTGAG | AtCCoAOMT1-QF | CTCAGGGAAGTGACAGCAAA | |
| AtPAL2-QR | TTCTCGGTTAGCGATTCACC | AtCCoAOMT1-QR | GTGGCGAGAAGAGAGTAGCC | |
| AtC4H-QF | ACTGGCTTCAAGTCGGAGAT | AtCCR1-QF | GTGCAAAGCAGATCTTCAGG | |
| AtC4H-QR | ACACGACGTTTCTCGTTCTG | AtCCR1-QR | GCCGCAGCATTAATTACAAA | |
| At4CL1-QF | TCAACCCGGTGAGATTTGTA TCGTCATCGATCAATCCAAT | AtF5H-QF | CTTCAACGTAGCGGATTTCA | |
| At4CL1-QR | TCGTCATCGATCAATCCAAT | AtF5H-QR | AGATCATTACGGGCCTTCAC |
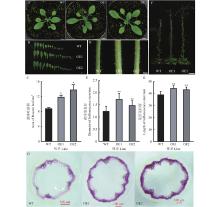
图6
过表达ClNAC40拟南芥的表型 A:4周龄野生型(WT)和过表达ClNAC40拟南芥植株(OE1、OE2)的表型;B,C:4周龄野生型(WT)和过表达ClNAC40拟南芥植株(OE1、OE2)的莲座叶数量及面积;D,E:4周龄野生型(WT)和过表达ClNAC40拟南芥植株(OE1、OE2)的花序茎直径;F,G:8周龄野生型(WT)和过表达ClNAC40拟南芥植株(OE1、OE2)的表型及花序茎高度;H:8周龄野生型与过表达ClNAC40拟南芥花序茎横切面;* 表示与WT相比存在显著差异(P<0.05);** 表示与WT相比存在极显著差异(P<0.01)。标尺Scale bars,A,B,D:1 cm;F:8 cm;H:100 μm。"

| 陈亚娟, 王宏芝, 李瑞芬, 等. 毛白杨纤维素合酶基因家族部分成员的克隆及表达. 林业科学, 2011, 47 (10): 70- 75. | |
| Chen Y J, Wang H Z, Li R F, et al. Isolation and expression profile of some members of cellulose synthase gene family in Populus tomentosa. Scientia Silvae Sinicae, 2011, 47 (10): 70- 75. | |
| 程健弘, 魏明科, 林二培, 等. 杉木HD-Zip Ⅲ 转录因子的克隆及表达分析. 农业生物技术学报, 2017, 25 (11): 1820- 1830. | |
| Cheng J H, Wei M K, Lin E P, et al. Cloning and expression analysis of HD-Zip Ш transcriptional factors in Cunninghamia lanceolata. Journal of Agricultural Biotechnology, 2017, 25 (11): 1820- 1830. | |
| 杜明秋, 钟珊丽, 林二培, 等. 2022. 杉木应压木形成中的显微特征及主要代谢成分变化. 核农学报, 36(11): 2307–2315. | |
| Du M Q, Zhong S L, Lin E P, et al. 2022. Anatomy characteristics and study of alterations of key metabolic components in Cunninghamia lanceolata during compression wood formation. Journal of Nuclear Agricultural Sciences, 36(11): 2307–2315. [in Chinese] | |
| 高文杰, 刘 娇, 马祥庆, 等. 杉木NAC基因家族基因的鉴定及生物信息学分析. 中南林业科技大学学报, 2022, 42 (2): 108- 118. | |
| Gao W J, Liu J, Ma X Q, et al. Identification and bioinformatics analysis of Chinese fir NAC gene family. Journal of Central South University of Forestry & Technology, 2022, 42 (2): 108- 118. | |
| 李 媛, 陈金焕, 金 曌, 等. 毛果杨NAC128基因在次生壁形成中的功能. 林业科学, 2020, 56 (11): 62- 72. | |
| Li Y, Chen J H, Jin Z, et al. Functions of NAC128 gene from Populus trichocarpa in secondary cell wall formation. Scientia Silvae Sinicae, 2020, 56 (11): 62- 72. | |
| 倪 飞, 励文豪, 林二培, 等. 光皮桦MYB基因的克隆及表达和调控分析. 林业科学, 2018, 54 (12): 70- 81. | |
| Ni F, Li W H, Lin E P, et al. Cloning, expression and regulation of MYB genes in Betula luminifera. Scientia Silvae Sinicae, 2018, 54 (12): 70- 81. | |
| 王 钰, 童再康, 黄华宏, 等. 光皮桦总RNA的提取及Actin基因的克隆. 浙江林业科技, 2010, 30 (4): 32- 36. | |
| Wang Y, Tong Z K, Huang H H, et al. Extracting of total RNA from Betula luminifera and the cloning of Actin gene. Journal of Zhejiang Forestry Science and Technology, 2010, 30 (4): 32- 36. | |
| 魏明科, 俞金健, 黄晓龙, 等. 杉木NAC转录因子基因ClNAC1的克隆、表达及单核苷酸多态性分析. 林业科学, 2018, 54 (9): 49- 59. | |
| Wei M K, Yu J J, Huang X L, et al. Cloning, expression and single nucleotide polymorphisms analysis of NAC transcription factor gene ClNAC1 in Cunninghamia lanceolata. Scientia Silvae Sinicae, 2018, 54 (9): 49- 59. | |
| 肖玉菲, 刘海龙, 刘雄盛, 等. 尾叶桉GLU4无性系F5H基因的克隆表达及序列分析. 生物技术, 2018, 28 (3): 205- 211. | |
| Xiao Y F, Liu H L, Liu X S, et al. Cloning, expression and sequence analysis of F5H gene in Eucalyptus urophylla clone GLU4. Biotechnology, 2018, 28 (3): 205- 211. | |
| 谢一青, 李志真, 黄儒珠, 等. 光皮桦基因组DNA提取方法比较. 浙江林学院学报, 2006, 23 (6): 664- 668. | |
| Xie Y Q, Li Z Z, Huang R Z, et al. Comparison of methods of extracting genomic DNA from Betula luminifera. Journal of Zhejiang A & F University, 2006, 23 (6): 664- 668. | |
| 徐莉莉, 童再康, 林二培, 等. 杉木苯丙氨酸解氨酶基因ClPAL的克隆与表达分析. 林业科学, 2013, 49 (12): 64- 72. | |
| Xu L L, Tong Z K, Lin E P, et al. Isolation and expression of CIPAL genes in Chinese fir (Cunninghamia lanceolata). Scientia Silvae Sinicae, 2013, 49 (12): 64- 72. | |
| 俞新妥. 中国杉木90年代的研究进展 Ⅰ. 杉木研究的特点及有关基础研究的综述. 福建林学院学报, 2000, 20 (1): 87- 96. | |
| Yu X T. A summary of the studies on Chinese fir in 1990's I. the distinguishing features of Chinese fir research and research development on basic research. Journal of Forest and Environment, 2000, 20 (1): 87- 96. | |
| Camargo E L, Ployet R, Cassan-Wang H, Mounet F, et al. Digging in wood: new insights in the regulation of wood formation in tree species. Advances in Botanical Research, 2019, 89, 201- 233. | |
|
Chen C J, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13 (8): 1194- 1202.
doi: 10.1016/j.molp.2020.06.009 |
|
|
Chen X H, Wang H T, Li X Y, et al. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4 (4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biology, 2019, 19, 231.
doi: 10.1186/s12870-019-1812-0 |
|
|
Huang H H, Xu L L, Tong Z K, et al. De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genomics, 2012, 13, 648.
doi: 10.1186/1471-2164-13-648 |
|
|
Hussey S G, Mizrachi E, Spokevicius A V, et al. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biology, 2011, 11, 173.
doi: 10.1186/1471-2229-11-173 |
|
|
Mitsuda N, Iwase A, Yamamoto H, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. The Plant Cell, 2007, 19 (1): 270- 280.
doi: 10.1105/tpc.106.047043 |
|
|
Ohtani M, Nishikubo N, Xu B, et al. A NAC domain protein family contributing to the regulation of wood formation in poplar. The Plant Journal, 2011, 67 (3): 499- 512.
doi: 10.1111/j.1365-313X.2011.04614.x |
|
| Sakamoto S, Mitsuda N. 2015. Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant and Cell Physiology, 56(2): 299–310. | |
| Salinas M, Xing S, Höhmann S, et al. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta, 2012, 235 (6): 1171- 1184. | |
|
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 2003, 13 (11): 2498- 2504.
doi: 10.1101/gr.1239303 |
|
|
Singh S, Koyama H, Bhati K K, et al. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. Journal of Plant Research, 2021, 134 (3): 475- 495.
doi: 10.1007/s10265-021-01270-y |
|
|
Sun Y M, Jiang C X, Jiang R Q, et al. A novel NAC transcription factor from Eucalyptus, EgNAC141, positively regulates lignin biosynthesis and increases lignin deposition. Frontiers in Plant Science, 2021, 12, 642090.
doi: 10.3389/fpls.2021.642090 |
|
| Taylor-Teeples M, Lin L, de Lucas M, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature, 2015, 517 (7536): 571- 575. | |
|
Wagner A, Tobimatsu Y, Phillips L, et al. CCoAOMT suppression modifies lignin composition in Pinus radiata. The Plant Journal, 2011, 67 (1): 119- 129.
doi: 10.1111/j.1365-313X.2011.04580.x |
|
| Wang H Z, Dixon R A. 2012. On-off switches for secondary cell wall biosynthesis. Molecular Plant, 5(2): 297–303. | |
|
Wang J F, Wang Y P, Zhang J, et al. The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Horticulture Research, 2021, 8 (1): 214.
doi: 10.1038/s41438-021-00649-1 |
|
| Zhang J, Serra J A, Helariutta Y. 2015. Wood development: growth through knowledge. Nature Plants, 1(5): 15060. | |
|
Zhang J, Xie M, Tuskan G A, et al. Recent advances in the transcriptional regulation of secondary cell wall biosynthesis in the woody plants. Frontiers in Plant Science, 2018, 9, 1535.
doi: 10.3389/fpls.2018.01535 |
|
|
Zhang X, Long Y, Chen X X, et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biology, 2021, 21 (1)
doi: 10.1186/s12870-020-02777-7 |
|
| Zhong R Q, Demura T, Ye Z H. 2006. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. The Plant Cell, 18(11): 3158–3170. | |
|
Zhong R Q, Lee C, Haghighat M, et al. Xylem vessel-specific SND5 and its homologs regulate secondary wall biosynthesis through activating secondary wall NAC binding elements. New Phytologist, 2021, 231 (4): 1496- 1509.
doi: 10.1111/nph.17425 |
|
|
Zhong R Q, Lee C, Zhou J L, et al. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. The Plant Cell, 2008, 20 (10): 2763- 2782.
doi: 10.1105/tpc.108.061325 |
|
|
Zhong R Q, Lee C, Ye Z H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends in Plant Science, 2010, 15 (11): 625- 632.
doi: 10.1016/j.tplants.2010.08.007 |
|
|
Zhong R Q, McCarthy R L, Lee C, et al. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiology, 2011, 157 (3): 1452- 1468.
doi: 10.1104/pp.111.181354 |
|
| Zhong R Q, Ye Z H. 2007. Regulation of cell wall biosynthesis. Current Opinion in Plant Biology, 10: 564–572. | |
| Zhong R Q, Ye Z H. 2015a. Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant and Cell Physiology, 56(2): 195–214. | |
| Zhong R Q, Ye Z H. 2015b. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signaling and Behavior, 10(2): e989746. | |
|
Zhuang H B, Chong S L, Priyanka B, et al. Full-length transcriptomic identification of R2R3-MYB family genes related to secondary cell wall development in Cunninghamia lanceolata (Chinese fir). BMC Plant Biology, 2021, 21 (1): 581.
doi: 10.1186/s12870-021-03322-w |
| [1] | 何江,覃林. 气候敏感的杉木天然林林分进界模型[J]. 林业科学, 2025, 61(1): 70-80. |
| [2] | 杨淑雅,王镜如,朱滢滢,伊力塔,刘美华. 杉木与浙江楠混交对根系分泌物和丛枝菌根真菌群落结构的影响[J]. 林业科学, 2024, 60(9): 59-68. |
| [3] | 都亚敏,李珠,蒋佳荔,殷方宇,吕建雄. 吸湿解吸循环过程中木材水分吸附特性[J]. 林业科学, 2024, 60(9): 150-158. |
| [4] | 李林鑫,杨贵云,郭昊澜,董强,李明,马祥庆,吴鹏飞. 繁殖方式对杉木幼苗根系不同序级生物量、形态性状和碳氮含量的影响[J]. 林业科学, 2024, 60(7): 47-55. |
| [5] | 阮颖超,苏比·热西塔洪,林熙,李明,范少辉,冯随起,陈志云,马祥庆,何宗明. 修枝强度对杉木人工林无节材形成及质量的影响[J]. 林业科学, 2024, 60(6): 50-59. |
| [6] | 吕梓晴, 段爱国. 不同产区杉木生物量与碳储量模型[J]. 林业科学, 2024, 60(2): 1-11. |
| [7] | 杨梦佳,邹显花,郭志娟,彭钊,何妍,彭志远,姚必达,黄国敏,朱丽琴,黄荣珍. 基于13C示踪的2个杉木家系幼苗光合碳分配动态[J]. 林业科学, 2024, 60(12): 35-46. |
| [8] | 贾辉,朱敏,余再鹏,万晓华,傅彦榕,王思荣,邹秉章,黄志群. 亚热带树种在未成林造林地的凋落物量和周转与叶片性状的关系[J]. 林业科学, 2024, 60(1): 12-18. |
| [9] | 李晓燕,段爱国,张建国. 不同产区杉木人工林初植密度对优势高生长的影响[J]. 林业科学, 2023, 59(8): 22-29. |
| [10] | 屈彦成,江怡航,姜彦妍,张建国,罗安利,张雄清. 基于胸高处边材面积、胸径和冠基部直径的杉木单木叶生物量预测模型[J]. 林业科学, 2023, 59(7): 106-114. |
| [11] | 赵文菲,曹小玉,谢政锠,庞一凡,孙亚萍,李际平,莫永俊,袁达. 基于结构方程模型的杉木公益林林分空间结构评价[J]. 林业科学, 2022, 58(8): 76-88. |
| [12] | 李舟阳,陆文玲,钱旺,黄奕孜,林二培,黄华宏,童再康. 杉木根边缘细胞生物学特性及其对铝胁迫的响应[J]. 林业科学, 2022, 58(7): 73-81. |
| [13] | 陈嘉琪,赵光宇,李仰龙,董玉红,厚凌宇,焦如珍. 杉木人工林土壤磷素形态及含量的林龄变化[J]. 林业科学, 2022, 58(5): 10-17. |
| [14] | 史月冬,郑宏,叶代全,施季森,边黎明. 杉木生长性状的空间与竞争效应及其对遗传参数估计的影响[J]. 林业科学, 2022, 58(5): 75-84. |
| [15] | 王淑真,梁晶晶,包明琢,潘菲,周垂帆. 不同林龄杉木林土壤磷形态与解磷菌变化[J]. 林业科学, 2022, 58(2): 58-69. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||