

林业科学 ›› 2024, Vol. 60 ›› Issue (1): 68-79.doi: 10.11707/j.1001-7488.LYKX20230138
张俊红1,王洋1,周生财2,吴小林3,吴仁超3,杨琪1,张毓婷1,童再康1,*( )
)
收稿日期:2023-04-06
出版日期:2024-01-25
发布日期:2024-01-29
通讯作者:
童再康
E-mail:zktong@zjfc.edu.cn
基金资助:
Junhong Zhang1,Yang Wang1,Shengcai Zhou2,Xiaolin Wu3,Renchao Wu3,Qi Yang1,Yuting Zhang1,Zaikang Tong1,*( )
)
Received:2023-04-06
Online:2024-01-25
Published:2024-01-29
Contact:
Zaikang Tong
E-mail:zktong@zjfc.edu.cn
摘要:
目的: 基于SSR分子标记技术研究珍稀濒危保护树种闽楠群体的遗传结构,构建核心种质资源库,为种质资源的科学管理、有效保护和高效利用提供理论依据。方法: 利用33对多态性SSR引物,分析来自福建、江西、湖南、浙江、广西5省(区)27个种源地218个闽楠家系425份种质资源群体的遗传多样性和遗传结构。应用DateTrans1.0联合Popgene32软件计算观测等位基因数(Na)、有效等位基因数(Ne)、观测杂合度(Ho)、期望杂合度(He)、Shannon信息指数(I) 和Nei’s基因多样性指数(H);运用STRUCTURE 2.3.4软件对9个闽楠群体进行遗传类群划分。采用最小距离逐步取样法构建核心种质库,通过对相关遗传参数的t检验验证核心种质库的有效性。结果: 依据种质来源地的地理分布,218个家系可分成9个群体;种质资源群体的平均有效等位基因数(Ne)、观测杂合度(Ho)、期望杂合度(He)均值、Shannon信息指数(I)分别为2.159、0.224、0.477和0.841,表明闽楠种质资源群体具较高遗传多样性;群体遗传结构分析表明,9个群体可划分为3个亚群;425份原始种质经最小距离逐步聚类取样得到85份核心种质和340份保留种质,核心种质占原始种质的20%,其Na、Ne、Ho、He、I和H保留率分别为92.318%、103.803%、116.652%、105.052%、103.341%和104.664%。t检验表明核心种质和原始种质的遗传多样性参数无显著差异,能充分代表原始种质的遗传多样性。结论: 构建的核心种质库在保留原始种质库遗传多样性的基础上,去除遗传冗余,有利于闽楠种质资源的有效保护和科学利用,为进一步育种工作奠定基础。
中图分类号:
张俊红,王洋,周生财,吴小林,吴仁超,杨琪,张毓婷,童再康. 闽楠群体遗传结构分析与核心种质库构建[J]. 林业科学, 2024, 60(1): 68-79.
Junhong Zhang,Yang Wang,Shengcai Zhou,Xiaolin Wu,Renchao Wu,Qi Yang,Yuting Zhang,Zaikang Tong. Genetic Structure Analysis and Core Germplasm Collection Construction of Phoebe bournei Populations[J]. Scientia Silvae Sinicae, 2024, 60(1): 68-79.
图1
闽楠天然种质资源的分布信息 ZH:政和Zhenghe;JO:建瓯Jian’ou;NP:南平Nanping;PC:浦城Pucheng;WYS:武夷山Wuyishan;SoX:松溪Songxi;SM:三明Sanming;YX:尤溪Youxi;ShX:沙县Shaxian;TN:泰宁Taining;JL:将乐Jiangle;MX:明溪Mingxi;YA:永安Yong’an;WP:武平Wuping;LS:龙胜Longsheng;XA:兴安Xing’an;SJ:三江Sanjiang;ShY:邵阳Shaoyang;YZ:永州Yongzhou;YL:炎陵Yanling;SR:上饶Shangrao;GZ:赣州Ganzhou;JA:吉安Ji’an;SC:遂川Suichuan;QY:庆元Qingyuan;SoY:松阳Songyang;TS:泰顺Taishun."
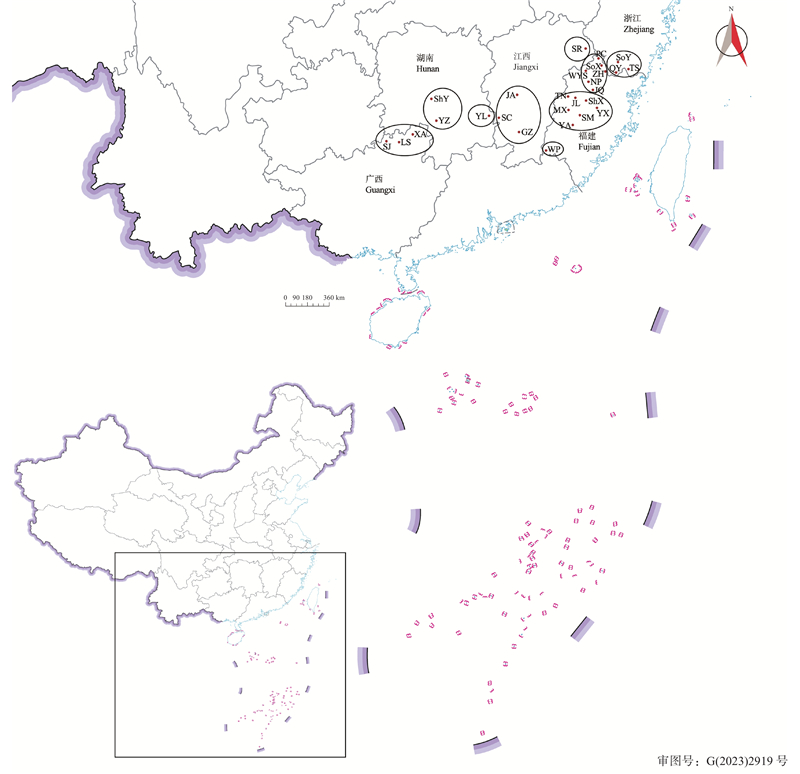
表1
闽楠种质资源信息"
| 群体 Population | 地理来源 Geographic source | 家系 Family | 种质数量 Sample size |
| 1 福建北部 Northern Fujian | 政和Zhenghe | 13, 14, 15, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88 | 18 |
| 建瓯Jian’ou | 10, 29, 30, 31, 32 | 5 | |
| 南平Nanping | 124, 203, 204, 205, 206, 207, 208 | 7 | |
| 浦城Pucheng | 67, 68, 69 | 3 | |
| 武夷山Wuyishan | 2, 89, 90, 91 | 4 | |
| 松溪Songxi | 3, 4, 12, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 118, 119, 121, 122, 123 | 26 | |
| 2 福建中部 Central Fujian | 三明Sanming | 5, 6, 7, 196, 197, 198 | 6 |
| 尤溪Youxi | 46 | 1 | |
| 沙县Shaxian | 8, 9, 71, 72, 73, 92, 93, 94, 95, 96, 97, 98 | 12 | |
| 泰宁Taining | 57, 58, 59, 60, 61, 62, 63, 64, 65, 66 | 10 | |
| 将乐Jiangle | 47, 48, 49, 50, 51, 52, 53, 54, 55, 56 | 10 | |
| 明溪Mingxi | 43, 44, 45 | 3 | |
| 永安Yong’an | 33, 34, 35, 36, 37, 38, 39, 40, 41, 42 | 10 | |
| 3 福建南部 Southern Fujian | 武平Wuping | 199, 200, 201, 202 | 4 |
| 4 广西北部 Northern Guangxi | 龙胜Longsheng | 21, 22, 229 | 3 |
| 兴安Xing’an | 24, 25, 26, 27, 28 | 5 | |
| 三江Sanjiang | 20 | 1 | |
| 5 湖南西部 Western Hunan | 邵阳Shaoyang | 184, 185, 186 | 3 |
| 永州Yongzhou | 192, 193, 194, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219, 220, 221, 222, 223, 224, 228 | 19 | |
| 6 湖南东部 Eastern Hunan | 炎陵Yanling | 181, 182, 183 | 3 |
| 7 江西北部 Northern Jiangxi | 上饶Shangrao | 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180 | 13 |
| 8 江西南部 Southern Jiangxi | 赣州Ganzhou | 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167 | 12 |
| 吉安Ji’an | 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155 | 23 | |
| 遂川Suichuan | 137, 138, 139, 140, 141, 142, 143, 144 | 8 | |
| 9 浙江南部 Southern Zhejiang | 庆元Qingyuan | 191, 225, 226, 227 | 4 |
| 松阳Songyang | 187, 188, 189, 190 | 4 | |
| 泰顺Taishun | 209 | 1 |
表2
33对SSR引物信息"
| 引物 Primer | 重复基元 Repeat motifs | 上游引物 Forward primer sequence (5’-3’) | 下游引物 Reverse primer sequence (5’-3’) | 预计产物长度 Size/bp |
| E-MN3 | (TGC)6 | TTGGTGGTGATAAGGGTGGT | TCAAAGGAAGAAAGCGCAAG | 154 |
| E-MN7 | (CAA)5 | GTAGGGAAGGTGTTGCTGGT | TCTCAAATGCAGATGCCGTA | 203 |
| E-MN8 | (GTT)6 | TTTCCACCACCCAAATTCAT | ATGGTCTCCAAGACGACGAC | 217 |
| E-MN11 | (ACACAA)4 | TAAGCAACGAATCCATGCAA | TCCTTCCATCTGCGTCTTTT | 178 |
| E-MN17 | (GGCTGG)4 | CCATGCGTCTGAGCAGATTA | GGTCCACTGATGCTGGCTAT | 241 |
| E-MN19 | (TG)7tatc(TATG)5 | TGGACATGCACAAACCATCT | GGCAAAGAAAGGCATTGAAG | 245 |
| E-MN21 | (AAT)6 | TCACCAGCCCCATCTTATTC | TGCCAACACTCATCCCATTA | 159 |
| E-MN25 | (AGTGC)4 | ATTCATCGAAGCTGGATTGG | CATCCCATCGTCCCAAATAC | 198 |
| E-MN35 | (TGT)7 | ATCCTCGTCCTCATCCTCCT | GAAATCATCGCAGAAGCACA | 250 |
| E-MN46 | (ATT)6 | TGTCGCGTGTCAAGCACTAT | CCATCTGCAATTCTCGTCAG | 225 |
| E-MN47 | (AAGGGA)4 | TTGTCGGGTCAGACTCAGTG | CTTTCCATTTCCCCCATTCT | 154 |
| E-MN54 | (TGA)6 | CCCCAACATGGTTAGGAAGA | TGTGGAACAGTGGAAGTGGA | 189 |
| E-MN58 | (TCT)5 | CCATCTCTCCACGATCCCTA | TAATTTCAACGAGGGCCAAG | 213 |
| E-MN62 | (AGA)5 | AACAGCGGAAAGAATCAACG | TTTCACTTCGGCTTCGTCTT | 229 |
| E-MN66 | (GGA)6 | CTTGAGGGTTTGTCGTTGGT | TGATGGAGTTTGCTGAGGTG | 213 |
| E-MN71 | (ACA)5 | GGTAAGGCCTCTCCTCTGCT | GTTACTCGGGCGATGACAAT | 203 |
| E-MN88 | (GCT)5 | GCAAACACTCTCCCACCATT | CCCACTCTTGTCTTGCTTCC | 202 |
| E-MN89 | (GAA)5 | GGATCGGAAGGTGAAGATGA | CTCCGAAACGTTCCTATCCA | 210 |
| E-MN90 | (GAA)5 | GAGAACCCATGGTTGGTTTG | TTGCACCCTCGATTCTCTCT | 208 |
| G-MN102 | (GATAAG)5 | TTGCATCGTTGAGGAGCTGT | CAGAGTCGCCAATGTCGTCT | 169 |
| G-MN114 | (CATT)5 | ATGCTGACCGTGGTTCGATT | CATGTGAGCCCATTCCAGGT | 208 |
| G-MN120 | (CCGGG)5 | GGTGTGTCCAAGAGGTGTGT | ACTACCAACCGTGTGCATGT | 260 |
| G-MN134 | (CATATC)5 | TGTGCTGTATTTGGTCGCCT | CGATGGCTGGAACTGTGGAT | 256 |
| G-MN140 | (TGGTTC)5 | TGACCTCGGGAAGACACTCT | ATCTAGTAGGCAGCATGCGC | 215 |
| G-MN145 | (AAGAGC)6 | GAGGGAGCCATCCTTGATCG | TCCCTTGAGGAGACCAGGTT | 198 |
| G-MN146 | (CCGAA)5 | TGCGGTGATGGAGAAAGCAT | TGACAGATCCAAGGCAGCTG | 195 |
| G-MN150 | (TCA)6 | GAGCGATCCAAAGCCAATGC | ACCAAACTGCTCAGGGATCG | 211 |
| G-MN157 | (AACCG)5 | TGAGACCGAAAGCTTGGGAC | ACAACATGCATTTGGTGGGC | 242 |
| G-MN161 | (GCAC)5 | CAGCTCCCACCTCAAAACCT | GCGTCTGACTTGCAACTGTG | 227 |
| G-MN168 | (ATGAGG)6 | GTTGAAGAAGTCGGGGGAGG | GACTCCATTTCGCATTCGGC | 165 |
| G-MN174 | (CTG)7 | ACACCATTACTGCTGCTGCT | AGTACAGCCCATTGTCACCG | 207 |
| G-MN186 | (CTCCAA)5 | GAGCCGCACTTCAACTAGGT | GGAGTGGGAGTTGGAGTTGG | 195 |
| G-MN191 | (TATCAC)5 | GATGGCAGCTTCAACTTGGC | TCCCCAACTAGGTGGGATGT | 237 |
表3
闽楠种质资源群体的遗传多样性①"
| 位点 Locus | Na | Ne | Ho | He | I | H | PIC |
| E-MN3 | 4.000 | 2.273 | 0.351 | 0.561 | 0.934 | 0.560 | 0.466 |
| E-MN7 | 3.000 | 2.417 | 0.002 | 0.587 | 0.986 | 0.586 | 0.521 |
| E-MN8 | 4.000 | 2.589 | 0.485 | 0.614 | 1.073 | 0.614 | 0.535 |
| E-MN11 | 3.000 | 1.646 | 0.181 | 0.393 | 0.663 | 0.392 | 0.337 |
| E-MN17 | 3.000 | 2.142 | 0.000 | 0.534 | 0.821 | 0.533 | 0.424 |
| E-MN19 | 3.000 | 2.306 | 0.647 | 0.567 | 0.908 | 0.566 | 0.470 |
| E-MN21 | 3.000 | 2.077 | 0.000 | 0.519 | 0.798 | 0.519 | 0.412 |
| E-MN25 | 4.000 | 2.980 | 0.238 | 0.665 | 1.202 | 0.664 | 0.601 |
| E-MN35 | 4.000 | 2.379 | 0.551 | 0.580 | 1.002 | 0.580 | 0.510 |
| E-MN46 | 4.000 | 2.320 | 0.320 | 0.570 | 0.935 | 0.569 | 0.474 |
| E-MN47 | 4.000 | 2.234 | 0.304 | 0.553 | 0.905 | 0.552 | 0.457 |
| E-MN54 | 3.000 | 2.166 | 0.054 | 0.539 | 0.897 | 0.538 | 0.466 |
| E-MN58 | 5.000 | 2.574 | 0.454 | 0.612 | 1.060 | 0.612 | 0.537 |
| E-MN62 | 3.000 | 2.950 | 0.358 | 0.662 | 1.090 | 0.661 | 0.587 |
| E-MN66 | 4.000 | 2.461 | 0.177 | 0.594 | 1.004 | 0.594 | 0.523 |
| E-MN71 | 3.000 | 2.996 | 0.424 | 0.667 | 1.098 | 0.666 | 0.592 |
| E-MN88 | 3.000 | 2.633 | 0.233 | 0.621 | 1.029 | 0.620 | 0.547 |
| E-MN89 | 3.000 | 1.977 | 0.318 | 0.495 | 0.738 | 0.494 | 0.385 |
| E-MN90 | 3.000 | 2.325 | 0.332 | 0.571 | 0.920 | 0.570 | 0.478 |
| G-MN102 | 4.000 | 1.329 | 0.000 | 0.248 | 0.543 | 0.248 | 0.237 |
| G-MN114 | 4.000 | 1.142 | 0.005 | 0.124 | 0.269 | 0.124 | 0.118 |
| G-MN120 | 2.000 | 1.200 | 0.000 | 0.167 | 0.307 | 0.167 | 0.153 |
| G-MN134 | 10.000 | 4.677 | 0.699 | 0.787 | 1.774 | 0.786 | 0.759 |
| G-MN140 | 3.000 | 1.437 | 0.000 | 0.305 | 0.562 | 0.304 | 0.275 |
| G-MN145 | 3.000 | 1.768 | 0.285 | 0.435 | 0.767 | 0.434 | 0.390 |
| G-MN146 | 4.000 | 1.643 | 0.009 | 0.392 | 0.765 | 0.391 | 0.361 |
| G-MN150 | 3.000 | 1.204 | 0.047 | 0.170 | 0.353 | 0.169 | 0.160 |
| G-MN157 | 2.000 | 1.978 | 0.000 | 0.495 | 0.688 | 0.494 | 0.372 |
| G-MN161 | 3.000 | 1.191 | 0.115 | 0.161 | 0.347 | 0.160 | 0.153 |
| G-MN168 | 3.000 | 1.158 | 0.000 | 0.136 | 0.284 | 0.136 | 0.129 |
| G-MN174 | 10.000 | 1.444 | 0.268 | 0.308 | 0.723 | 0.307 | 0.295 |
| G-MN186 | 9.000 | 4.090 | 0.355 | 0.756 | 1.623 | 0.756 | 0.718 |
| G-MN191 | 4.000 | 1.532 | 0.169 | 0.348 | 0.678 | 0.347 | 0.319 |
| 平均值Mean | 3.939 | 2.159 | 0.224 | 0.477 | 0.841 | 0.476 | 0.417 |
表4
9个闽楠种质资源群体的遗传多样性①"
| 群体Population | Na | Ne | Ho | He | H | I |
| 福建北部 Northern Fujian | 3.636 | 2.049 | 0.227 | 0.462 | 0.460 | 0.797 |
| 福建南部Southern Fujian | 2.455 | 1.968 | 0.280 | 0.474 | 0.444 | 0.713 |
| 福建中部Central Fujian | 3.394 | 1.912 | 0.233 | 0.423 | 0.421 | 0.723 |
| 广西北部Northern Guangxi | 2.758 | 1.830 | 0.157 | 0.421 | 0.409 | 0.692 |
| 湖南东部Eastern Hunan | 2.364 | 1.924 | 0.263 | 0.408 | 0.374 | 0.623 |
| 湖南西部Western Hunan | 3.455 | 2.287 | 0.248 | 0.516 | 0.510 | 0.892 |
| 江西北部Northern Jiangxi | 2.242 | 1.621 | 0.218 | 0.313 | 0.306 | 0.504 |
| 江西南部Southern Jiangxi | 2.788 | 1.957 | 0.201 | 0.406 | 0.404 | 0.674 |
| 浙江南部Southern Zhejiang | 3.000 | 2.155 | 0.226 | 0.471 | 0.458 | 0.786 |
| 平均值Mean | 2.899 | 1.967 | 0.228 | 0.433 | 0.421 | 0.712 |
表5
各取样比例下的遗传多样性比较①"
| 取样比例 Sampling ratio | 种质数目 Germplasm number | Na | Ne | Ho | He | I | H |
| 50 | 213 | 3.849 | 2.093 | 0.242 | 0.461 | 0.805 | 0.459 |
| 45 | 191 | 3.849 | 2.102 | 0.245 | 0.463 | 0.809 | 0.461 |
| 40 | 170 | 3.849 | 2.123 | 0.245 | 0.467 | 0.818 | 0.466 |
| 35 | 149 | 3.849 | 2.141 | 0.249 | 0.475 | 0.833 | 0.474 |
| 30 | 128 | 3.788 | 2.172 | 0.247 | 0.482 | 0.844 | 0.480 |
| 25 | 106 | 3.758 | 2.198 | 0.254 | 0.491 | 0.858 | 0.489 |
| 20 | 85 | 3.636 | 2.241 | 0.261 | 0.501 | 0.869 | 0.498 |
| 15 | 64 | 3.606 | 2.168 | 0.268 | 0.491 | 0.846 | 0.487 |
| 10 | 43 | 3.606 | 2.204 | 0.277 | 0.497 | 0.857 | 0.491 |
表6
原始种质、核心种质和保留种质的遗传多样性比较①"
| 群体 Population | Na | Ne | Ho | He | I | H |
| 原始种质Original germplasms | 3.939 | 2.159 | 0.224 | 0.477 | 0.841 | 0.476 |
| 核心种质Core germplasms | 3.636 | 2.241 | 0.261 | 0.501 | 0.869 | 0.498 |
| 保留率Retention rate(%) | 92.318 | 103.803 | 116.652 | 105.052 | 103.341 | 104.664 |
| 保留种质Reserved germplasms | 3.909 | 2.122 | 0.215 | 0.465 | 0.819 | 0.465 |
| 保留率Retention rate(%) | 99.241 | 98.300 | 95.804 | 97.568 | 97.360 | 97.647 |
表7
核心种质与原始种质的遗传多样性参数t检验①"
| 参数 Parameters | 群体 Population | 均值 Mean | 标准差 Standard deviation | 均值差值 Mean difference | t | P |
| Na | 原始种质Original germplasms | 3.939 | 1.952 | 0.303 | 0.690 | 0.493 |
| 核心种质Core germplasms | 3.636 | 1.597 | ||||
| Ne | 原始种质Original germplasms | 2.159 | 0.803 | ?0.082 | ?0.412 | 0.681 |
| 核心种质Core germplasms | 2.241 | 0.821 | ||||
| Ho | 原始种质Original germplasms | 0.224 | 0.205 | ?0.038 | ?0.716 | 0.477 |
| 核心种质Core germplasms | 0.261 | 0.223 | ||||
| He | 原始种质Original germplasms | 0.477 | 0.185 | ?0.024 | ?0.535 | 0.595 |
| 核心种质Core germplasms | 0.501 | 0.185 | ||||
| I | 原始种质Original germplasms | 0.841 | 0.339 | ?0.028 | ?0.329 | 0.743 |
| 核心种质Core germplasms | 0.869 | 0.363 | ||||
| H | 原始种质Original germplasms | 0.476 | 0.185 | ?0.022 | ?0.484 | 0.630 |
| 核心种质Core germplasms | 0.498 | 0.184 |
表8
闽楠核心种质分布情况"
| 类群 Group | 分布区域 Distribution area | 地理来源 Geographic source | 家系 Family |
| 第一类群 (蓝色部分) The first group (blue part) | 1 福建北部Northern Fujian | 政和 Zhenghe | 13, 87 |
| 建瓯Jian’ou | 29, 31 | ||
| 南平Nanping | 203, 204, 206, 207, 208 | ||
| 松溪Songxi | 3, 100, 101, 102, 103, 104, 105, 106, 107, 108, 111, 112, 114, 115, 116, 118, 119, 122 | ||
| 2 福建中部Central Fujian | 三明Sanming | 5, 7, 196(2), 197, 198 | |
| 沙县Shaxian | 9, 73, 94, 97 | ||
| 将乐Jiangle | 51 | ||
| 明溪Mingxi | 43 | ||
| 第二类群 (绿色部分) The second group (green part) | 3 福建南部Southern Fujian | 武平Wuping | 200, 201, 202 |
| 5 湖南西部Western Hunan | 邵阳Shaoyang | 184, 186 | |
| 永州Yongzhou | 192, 193, 210, 211, 212, 213, 215, 216, 217, 218(2), 219, 220, 222, 223(2), 228 | ||
| 6 湖南东部Eastern Hunan | 炎陵Yanling | 181, 182, 183(2) | |
| 8 江西南部Southern Jiangxi | 吉安Ji’an | 130, 147, 151 | |
| 遂川Suichuan | 138, 141 | ||
| 9 浙江南部Southern Zhejiang | 庆元Qingyuan | 191(2), 225, 226, 227(2) | |
| 松阳Songyang | 187, 188(2), 190 | ||
| 泰顺Taishun | 209 | ||
| 第三群类 (红色部分) The third group (red part) | 4 广西北部Northern Guangxi | 兴安Xing’an | 26 |
| 三江Sanjiang | 20 | ||
| 7 江西北部Northern Jiangxi | 上饶Shangrao | 168, 172 |
|
陈 存, 丁昌俊, 张 静, 等. 美洲黑杨群体结构分析及核心种质库构建. 林业科学, 2020, 56 (9): 67- 76.
doi: 10.11707/j.1001-7488.20200908 |
|
|
Chen C, Ding C J, Zhang J, et al. Population structure analysis and core collection construction of Populus deltoides. Scientia Silvae Sinicae, 2020, 56 (9): 67- 76.
doi: 10.11707/j.1001-7488.20200908 |
|
| 冯一宁, 李因刚, 祁 铭, 等. 基于SSR标记的福建省闽楠代表性群体遗传多样性分析. 南京林业大学学报(自然科学版), 2022, 46 (4): 102- 108. | |
| Feng Y N, Li Y G, Qi M, et al. Genetic diversity analyses of Phoebe bournei representative populations in Fujian Province based on SSR markers. Journal of Nanjing Forestry University (Natural Sciences Edition), 2022, 46 (4): 102- 108. | |
| 黄雨芹, 尹光天, 杨锦昌, 等. 基于SSR分子标记的闽楠(Phoebe bournei)核心种质的构建. 分子植物育种, 2020, 18 (8): 2641- 2648. | |
| Huang Y Q, Yin G T, Yang J C, et al. Developing a mini core germplasm of Phoebe bournei based on SSR molecular marker. Molecular Plant Breeding, 2020, 18 (8): 2641- 2648. | |
|
李洪果, 许基煌, 杜红岩, 等. 基于等位基因最大化法初步构建杜仲核心种质. 林业科学, 2018, 54 (2): 42- 51.
doi: 10.11707/j.1001-7488.20180205 |
|
|
Li H G, Xu J H, Du H Y, et al. Preliminary construction of core collection of Eucommia ulmoides based on allele number maximization strategy. Scientia Silvae Sinicae, 2018, 54 (2): 42- 51.
doi: 10.11707/j.1001-7488.20180205 |
|
|
李魁鹏, 陈仕昌, 程 琳, 等. 基于SSR标记构建广西杉木核心种质. 广西科学, 2021, 28 (5): 511- 519.
doi: 10.13656/j.cnki.gxkx.20211203.003 |
|
|
Li K P, Chen S C, Cheng L, et al. Construction of core germplasm of Cunninghamia lanceolata in Guangxi based on SSR markers. Guangxi Sciences, 2021, 28 (5): 511- 519.
doi: 10.13656/j.cnki.gxkx.20211203.003 |
|
| 刘 丹. 2019. 闽楠种质资源遗传多样性的SSR分析. 福州: 福建农林大学. | |
| Liu D. 2019. Genetic diversity of germplasm resources revealed by SSR in Phoebe bourne. Fuzhou: Fujian Agriculture and Forestry University. [in Chinese] | |
| 刘 军, 姜景民, 陈益泰, 等. 闽楠种子轻基质容器育苗及优良家系选择. 西北林学院学报, 2011, 26 (6): 70- 73,228. | |
| Liu J, Jiang J M, Chen Y T, et al. Cultural techniques of container seedlings with light medium and family selection for Phoebe bournei. Journal of Northwest Forestry University, 2011, 26 (6): 70- 73,228. | |
|
明 军, 张启翔, 兰彦平. 梅花品种资源核心种质构建. 北京林业大学学报, 2005, 27 (2): 65- 69.
doi: 10.3321/j.issn:1000-1522.2005.02.013 |
|
|
Ming J, Zhang Q X, Lan Y P. Core collection of Prunus mume Sieb. et Zucc. Journal of Beijing Forestry University, 2005, 27 (2): 65- 69.
doi: 10.3321/j.issn:1000-1522.2005.02.013 |
|
|
潘英华, 徐志健, 梁云涛. 广西普通野生稻群体结构解析与核心种质构建. 植物遗传资源学报, 2018, 19 (3): 498- 509.
doi: 10.13430/j.cnki.jpgr.2018.03.015 |
|
|
Pan Y H, Xu Z J, Liang Y T. Genetic structure and core collection of common wild rice (Oryza rufipogon Griff. ) in Guangxi. Journal of Plant Genetic Resources, 2018, 19 (3): 498- 509.
doi: 10.13430/j.cnki.jpgr.2018.03.015 |
|
|
申 展, 马履一, 敖 妍, 等. 基于SSR标记的文冠果遗传多样性分析及核心种质构建. 分子植物育种, 2017, 15 (8): 3341- 3350.
doi: 10.13271/j.mpb.015.003341 |
|
|
Shen Z, Ma L Y, Ao Y, et al. Analysis of the genetic diversity and construction of core collection of Xanthoceras sorbifolia Bunge. using microsatellite marker data. Molecular Plant Breeding, 2017, 15 (8): 3341- 3350.
doi: 10.13271/j.mpb.015.003341 |
|
| 孙荣喜. 2017. 中国枫香树遗传多样性及谱系地理研究. 北京: 中国林业研究科学院. | |
| Sun R X. 2017. Genetic diversity and phylogeography of Liquidambar formosana Hance in China. Beijing: Chinese Academy of Forestry. [in Chinese] | |
|
王 雪, 高 暝, 吴立文, 等. 峨眉山地区杨叶木姜子群体遗传多样性研究. 植物遗传资源学报, 2019, 20 (2): 359- 369.
doi: 10.13430/j.cnki.jpgr.20180812001 |
|
|
Wang X, Gao M, Wu L W, et al. A study on population genetic diversity among Litsea populifolia (Hemsl.) gamble in mount Emei area. Journal of Plant Genetic Resources, 2019, 20 (2): 359- 369.
doi: 10.13430/j.cnki.jpgr.20180812001 |
|
| 吴大荣, 王伯荪. 濒危树种闽楠种子和幼苗生态学研究(英文). 生态学报, 2011, 21 (11): 1751- 1760. | |
| Wu D R, Wang B S. Seed and seedling ecology of the endangered Phoebe bournei (Lauraceae). Acta Ecologica Sinica, 2011, 21 (11): 1751- 1760. | |
|
吴大荣, 朱政德. 福建省罗卜岩自然保护区闽楠种群结构和空间分布格局初步研究. 林业科学, 2003, 39 (1): 23- 30.
doi: 10.3321/j.issn:1001-7488.2003.01.004 |
|
|
Wu D R, Zhu Z D. Preliminary study on structure and spatial distribution pattern of Phoebe bournei in Luo Boyan nature reserve in Fujian Province. Scientia Silvae Sinicae, 2003, 39 (1): 23- 30.
doi: 10.3321/j.issn:1001-7488.2003.01.004 |
|
|
徐海明, 邱英雄, 胡 晋, 等. 不同遗传距离聚类和抽样方法构建作物核心种质的比较. 作物学报, 2004, 30 (9): 932- 936.
doi: 10.3321/j.issn:0496-3490.2004.09.016 |
|
|
Xu H M, Qiu Y X, Hu J, et al. Methods of constructing core collection of crop germplasm by comparing different genetic distances, cluster methods and sampling strategies. Acta Agronomica Sinica, 2004, 30 (9): 932- 936.
doi: 10.3321/j.issn:0496-3490.2004.09.016 |
|
|
杨汉波, 张 蕊, 王帮顺, 等. 基于SSR标记的木荷核心种质构建. 林业科学, 2017, 53 (6): 37- 46.
doi: 10.11707/j.1001-7488.20170605 |
|
|
Yang H B, Zhang R, Wang B S, et al. Construction of core collection of Schima superba based on SSR molecular markers. Scientia Silvae Sinicae, 2017, 53 (6): 37- 46.
doi: 10.11707/j.1001-7488.20170605 |
|
|
杨欣超, 张凯权, 王 静, 等. 基于SSR分子标记的刺槐遗传多样性分析及核心种质的构建. 分子植物育种, 2020, 18 (9): 3086- 3097.
doi: 10.13271/j.mpb.018.003086 |
|
|
Yang X C, Zhang K Q, Wang J, et al. Analysis of genetic diversity and construction of core collections of black locust based on SSR markers. Molecular Plant Breeding, 2020, 18 (9): 3086- 3097.
doi: 10.13271/j.mpb.018.003086 |
|
|
张春雨, 陈学森, 张艳敏, 等. 采用分子标记构建新疆野苹果核心种质的方法. 中国农业科学, 2009, 42 (2): 597- 604.
doi: 10.3864/j.issn.0578-1752.2009.02.026 |
|
|
Zhang C Y, Chen X S, Zhang Y M, et al. A method for constructing core collection of Malus sieversii using molecular markers. Scientia Agricultura Sinica, 2009, 42 (2): 597- 604.
doi: 10.3864/j.issn.0578-1752.2009.02.026 |
|
|
张俊红, 黄华宏, 童再康, 等. 光皮桦6个南方天然群体的遗传多样性. 生物多样性, 2010, 18 (3): 233- 240.
doi: 10.3724/SP.J.1003.2010.233 |
|
|
Zhang J H, Huang H H, Tong Z K, et al. Genetic diversity in six natural populations of Betula luminifera from southern China. Biodiversity Science, 2010, 18 (3): 233- 240.
doi: 10.3724/SP.J.1003.2010.233 |
|
|
郑 健, 郑勇奇, 张川红, 等. 花楸树天然群体的遗传多样性研究. 生物多样性, 2008, 16 (6): 562- 569.
doi: 10.3321/j.issn:1005-0094.2008.06.006 |
|
|
Zheng J, Zheng Y Q, Zhang C H, et al. Genetic diversity in natural populations of Sorbus pohuashanensis. Biodiversity Science, 2008, 16 (6): 562- 569.
doi: 10.3321/j.issn:1005-0094.2008.06.006 |
|
|
Belaj A, Rosa R, Lorite I J, et al. Usefulness of a new large set of high throughput EST-SNP markers as a tool for olive germplasm collection management. Frontiers in Plant Science, 2018, 9, 1320.
doi: 10.3389/fpls.2018.01320 |
|
| Brown A. 1989. Core collections: a practical approach to genetic resource management. Genome, 31: 818-824. | |
|
Castelán P M, Cortés-Cruz M, Mendoza-Castillo M D C, et al. Diversity and genetic structure inferred with microsatellites in natural populations of Pseudotsuga menziesii (Mirb.) Franco (Pinaceae) in the central region of Mexico. Forests, 2019, 10 (2): 101.
doi: 10.3390/f10020101 |
|
| Charters Y M, Robertson A, Wilkinson M J, et al. PCR analysis of oilseed rape cultivars (Brassica napus L. ssp. oleifera) using 5'-anchored simple sequence repeat (SSR) primers. Theoretical & Applied Genetics, 1996, 92 (3): 442- 447. | |
|
Ding Y J, Zhang J H, Lu Y F, et al. Development of EST-SSR markers and analysis of genetic diversity in natural populations of endemic and endangered plant Phoebe chekiangensis. Biochemical Systematics and Ecology, 2015, 63, 183- 189.
doi: 10.1016/j.bse.2015.10.008 |
|
| Diwan N, Mcintosh M S, Bauchan G R. Methods of developing a core collection of annual Medicago species. Theoretical and Applied Genetics, 1995, 90 (5): 755- 761. | |
| Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry, 1987, 19 (1): 11- 13. | |
| Frankel O H, Brown A. 1984. Plant genetic resources today: a critical appraisal//Holden J H W, Williams J T. eds. Crop genetic resources: conservation and evaluation. London: George Allen and Unwin, 249–257. | |
| Gresshoff P M. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Analytical Biochemistry, 196(1): 80–83. | |
| Hamrick J L. 1989. Allozyme diversity in plant species//Brown H D, Clegg M T, Kahler A L, et al. eds. Plant population genetics, breeding, and genetic resources. Sunderland: Sinauer Associates, 43–63. | |
| Hamrick J L, Godt M J W, Sherman-Broyles S L. Factors influencing levels of genetic diversity in woody plant species. New Forests, 1992, 6 (1): 95- 124. | |
|
Han X, Zhang J H, Han S, et al. The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Communications, 2022, 3 (6): 100410.
doi: 10.1016/j.xplc.2022.100410 |
|
| Hu J, Zhu J, Xu H M. Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theoretical & Applied Genetics, 2000, 101 (1): 264- 268. | |
|
Kang C W, Kim S Y, Lee S W, et al. Selection of a core collection of Korean sesame germplasm by a stepwise clustering method. Breeding Science, 2006, 56 (1): 85- 91.
doi: 10.1270/jsbbs.56.85 |
|
|
Pearse D E, Crandall K A. Beyond FST: analysis of population genetic data for conservation. Conservation Genetics, 2004, 5 (5): 585- 602.
doi: 10.1007/s10592-003-1863-4 |
|
|
Rubio-Moraga A, Candel-Perez D, Lucas-Borja M E, et al. Genetic diversity of Pinus nigra Arn. populations in southern Spain and northern Morocco revealed by inter-simple sequence repeat profiles. International Journal of Molecular Sciences, 2012, 13 (5): 5645- 5658.
doi: 10.3390/ijms13055645 |
|
|
Shi X M, Qiang W, Mu C, et al. Genetic diversity and structure of natural Quercus variabilis population in China as revealed by microsatellites markers. Forests, 2017, 8 (12): 495.
doi: 10.3390/f8120495 |
|
| Wang H X, Zhao S G, Gao Y, et al. A construction of the core-collection of Juglans regia L. based on AFLP molecular markers. Scientia Agricultura Sinica, 2013, 46 (23): 4985- 4995. | |
|
Wang P L, Su J X, Wu H Y, et al. Analysis of germplasm genetic diversity and construction of a core collection in Camellia oleifera C.Abel by integrating novel simple sequence repeat markers. Genetic Resources Crop Evolution, 2023, 70 (5): 1517- 1530.
doi: 10.1007/s10722-022-01519-y |
|
|
Wang Y, Ma X, Lu Y, et al. Assessing the current genetic structure of 21 remnant populations and predicting the impacts of climate change on the geographic distribution of Phoebe sheareri in southern China. Science of the Total Environment, 2022, 846, 157391.
doi: 10.1016/j.scitotenv.2022.157391 |
|
| Wright S. Evolution in Mendelian populations. Genetics, 1931, 16 (2): 97- 159. | |
|
Wuyun T N, Amo H, Xu J S, et al. Population structure of and conservation strategies for wild Pyrus ussuriensis maxim in China. PLoS One, 2015, 10 (8): e0133686.
doi: 10.1371/journal.pone.0133686 |
|
| Yeh F C, Yang R C, Boyle T. 2000. PopGene32, Microsoft windows-based freeware for population genetic analysis, Version 1.32. | |
|
Zhou Q, Zhou P Y, Zou W T, et al. EST-SSR marker development based on transcriptome sequencing and genetic analyses of Phoebe bournei (Lauraceae). Molecular Biology Reports, 2021, 48 (3): 2201- 2208.
doi: 10.1007/s11033-021-06228-w |
|
|
Zhu Y Z, Liang D Y, Song Z J, et al. Genetic diversity analysis and core germplasm collection construction of Camellia oleifera based on fruit phenotype and SSR data. Genes (Basel), 2022, 13 (12): 2351.
doi: 10.3390/genes13122351 |
| [1] | 丁晓磊,汪青桐,林司曦,赵瑞文,张悦,叶建仁. 广东省与江苏省松材线虫种群遗传结构差异分析[J]. 林业科学, 2022, 58(8): 1-9. |
| [2] | 王妍,冯金玲,吴小慧,黄蓝明,吴娟,陈宇,杨志坚. 施肥对闽楠幼苗光合碳固定的影响[J]. 林业科学, 2022, 58(5): 40-52. |
| [3] | 潘文婷,孙建军,原勤勤,张利利,邓康桥,厉月桥. RAD-seq技术研究鹅掌楸属种源遗传多样性和遗传结构[J]. 林业科学, 2022, 58(4): 74-81. |
| [4] | 张苗,周生财,吴梦洁,童再康,韩潇,张俊红,程龙军. 闽楠中WRKY家族成员鉴定及缺磷胁迫下表达分析[J]. 林业科学, 2022, 58(2): 133-147. |
| [5] | 杜超群,孙晓梅,谢允慧,侯义梅. 北亚热带日本落叶松不同改良水平群体的遗传多样性[J]. 林业科学, 2021, 57(5): 68-76. |
| [6] | 王晓,韦小丽,吴高殷,陈胜群. CO2浓度升高条件下不同氮素供应对闽楠幼苗光合特性及生长的影响[J]. 林业科学, 2021, 57(4): 173-181. |
| [7] | 陈存,丁昌俊,张静,李波,褚延广,苏晓华,黄秦军. 美洲黑杨群体结构分析及核心种质库构建[J]. 林业科学, 2020, 56(9): 67-76. |
| [8] | 李霞,王利宝,文亚峰,林军,武星彤,袁美灵,张原,王敏求,李鑫玉. 杉木不同世代育种群体的遗传多样性[J]. 林业科学, 2020, 56(11): 53-61. |
| [9] | 马松梅, 王春成, 孙芳芳, 魏博, 聂迎彬. 濒危植物新疆野扁桃的遗传多样性[J]. 林业科学, 2019, 55(9): 71-80. |
| [10] | 金玲, 刘明国, 董胜君, 吴月亮, 张欣. 97个山杏无性系的遗传多样性及SSR指纹图谱[J]. 林业科学, 2018, 54(7): 51-61. |
| [11] | 包文泉, 乌云塔娜, 杜红岩, 李铁柱, 刘慧敏, 王淋, 白玉娥. 基于SSR标记的西藏光核桃群体遗传多样性和遗传结构分析[J]. 林业科学, 2018, 54(2): 30-41. |
| [12] | 杨汉波, 张蕊, 王帮顺, 徐肇友, 陈焕伟, 周志春. 木荷优树无性系种质SSR标记的遗传多样性分析[J]. 林业科学, 2017, 53(5): 43-53. |
| [13] | 毛秀红, 郑勇奇, 孙百友, 张元帅, 韩丛聪, 位晓, 荀守华. 基于SSR的刺槐无性系遗传多样性分析和指纹图谱构建[J]. 林业科学, 2017, 53(10): 80-89. |
| [14] | 冯源恒, 李火根, 杨章旗, 黄永利, 罗群凤, 张远. 广西马尾松第2代育种群体的组建[J]. 林业科学, 2017, 53(1): 54-61. |
| [15] | 宋志姣, 杨合宇, 翁启杰, 周长品, 李发根, 李梅, 卢万鸿, 罗建中, 甘四明. 细叶桉群体的遗传多样性和受选择位点[J]. 林业科学, 2016, 52(9): 39-47. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||