

Scientia Silvae Sinicae ›› 2026, Vol. 62 ›› Issue (1): 32-41.doi: 10.11707/j.1001-7488.LYKX20250221
• Frontiers and hot topics • Previous Articles Next Articles
Chenchen Guo1,Qi Li1,Siyuan Li1,Zemin Wang2,Yingnan Chen1,*( ),Suyun Wei1,Jianjun Hu3,*(
),Suyun Wei1,Jianjun Hu3,*( )
)
Received:2025-04-11
Revised:2025-10-14
Online:2026-01-25
Published:2026-01-14
Contact:
Yingnan Chen,Jianjun Hu
E-mail:chenyingnan@njfu.edu.cn;hujj@caf.ac.cn
CLC Number:
Chenchen Guo,Qi Li,Siyuan Li,Zemin Wang,Yingnan Chen,Suyun Wei,Jianjun Hu. Genomic Selection for Dynamic Growth Traits during the Seedling Stage of Poplar Hybrid Population[J]. Scientia Silvae Sinicae, 2026, 62(1): 32-41.
Table 1
Statistical analysis of phenotypic traits in hybrid population"
| 性状 Traits | 月份 Month | 最小值 Min. | 最大值 Max. | 均值 Mean | 变异系数 CV | 正态性检验# Norm test | 遗传力 Heritability |
| GD/mm | 4 | 0.75 | 7.25 | 3.65 | 0.34 | 0.98 | 0.42 |
| 5 | 1.95 | 8.01 | 4.55 | 0.28 | 0.99 | 0.42 | |
| 6 | 2.46 | 17.93 | 10.47 | 0.25 | 0.99 | 0.47 | |
| 7 | 3.31 | 27.15 | 16.74 | 0.25 | 0.99 | 0.42 | |
| 8 | 6.11 | 38.75 | 23.90 | 0.24 | 0.99 | 0.45 | |
| 9 | 7.00 | 49.23 | 29.01 | 0.23 | 0.98 | 0.42 | |
| 12 | 7.15 | 53.16 | 32.24 | 0.23 | 0.99 | 0.51 | |
| PH/cm | 4 | 6.10 | 126.90 | 46.85 | 0.54 | 0.95 | 0.43 |
| 5 | 8.50 | 154.30 | 54.91 | 0.48 | 0.94 | 0.39 | |
| 6 | 12.50 | 169.40 | 90.33 | 0.27 | 0.99 | 0.53 | |
| 7 | 15.00 | 205.40 | 127.77 | 0.23 | 0.98 | 0.60 | |
| 8 | 51.50 | 279.00 | 186.47 | 0.19 | 0.99 | 0.58 | |
| 9 | 65.00 | 400.00 | 279.02 | 0.18 | 0.97 | 0.62 | |
| 12 | 70.00 | 439.00 | 288.04 | 0.18 | 0.97 | 0.63 |
Table 3
Statistical analysis of observed values of growth traits in December and model predicted breeding values of BayesA model in September"
| 性状 Trait | 模型 Model | 最小值 Min. | 最大值 Max. | 均值 Mean | 方差 SD |
| GD/mm | OBV | 7.15 | 53.16 | 32.24 | 7.50 |
| BayesA | 22.06 | 37.81 | 29.19 | 2.56 | |
| PH/cm | OBV | 70.00 | 439.00 | 288.04 | 52.86 |
| BayesA | 202.24 | 335.37 | 278.95 | 22.39 |

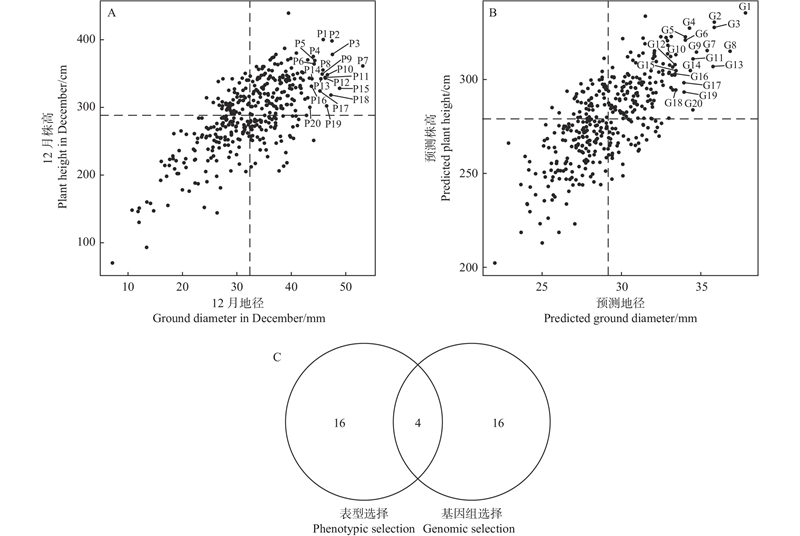
Fig.4
Comparative analysis of phenotypic selection and genomic selection A. Selection of elite individuals based on phenotypic data;B. Selection of elite individuals based on the BayesA model;C. Venn diagram comparing phenotypic selection and genomic selection of elite individuals.P1: 10-86-1; P2: 10-82-2; P3: 9-77-2; P4: 9-78-1; P5: 10-87-1; P6: 10-81-2; P7: 9-86-3; P8: 9-80-1; P9: 10-23-1; P10: 9-84-2; P11: 9-68-2; P12: 10-66-2; P13: 9-76-3; P14: 9-94-2; P15: 10-92-1; P16: 10-77-1; P17: 10-46-3; P18: 9-10-2; P19: 10-94-1; P20: 9-29-2; G1: 9-68-2; G2: 10-36-3; G3: 10-69-3; G4: 10-81-1; G5: 10-66-1; G6: 9-17-3; G7: 9-29-2; G8: 9-90-2; G9: 10-44-2; G10: 10-40-1; G11: 10-77-1; G12: 10-57-3; G13: 9-86-3; G14: 9-16-1; G15: 9-91-3; G16: 9-90-3; G17: 10-43-2; G18: 10-35-2; G19: 9-27-2; G20: 9-75-2."
| 杜常健, 张 敏, 周星鲁, 等. 杨树杂交群体苗期生长性状的全基因组选择研究. 林业科学研究, 2023, 36 (6): 11- 19. | |
| Du C J, Zhang M, Zhou X L, et al. Genome-wide selection of seedling growth traits in poplar hybrid populations. Forest Research, 2023, 36 (6): 11- 19. | |
| 杜庆章, 战鹏宇, 李 鹏, 等. 基因组选择研究进展及其在林木中的发展趋势. 北京林业大学学报, 2020, 42 (11): 1- 8. | |
| Du Q Z, Zhan P Y, Li P, et al. Advances in genomic selection and its development trend in forest. Journal of Beijing Forestry University, 2020, 42 (11): 1- 8. | |
| 贾会霞, 孙 佩, 李建波, 等. 丹红杨×转BtCry1Ac欧洲黑杨杂交子代抗虫性及生长量测定. 分子植物育种, 2017, 15 (10): 4101- 4109. | |
| Jia H X, Sun P, Li J B, et al. Insect-resistance and growth measurement of hybrid progeny from Populus deltoides cl. Danhong and transgenic P. nigra with BtCry1Ac gene. Molecular Plant Breeding, 2017, 15 (10): 4101- 4109. | |
| 康向阳. 论林木常规育种与非常规育种及其关系. 北京林业大学学报, 2023, 45 (6): 1- 7. | |
| Kang X Y. On conventional and unconventional tree breeding and their relationships. Journal of Beijing Forestry University, 2023, 45 (6): 1- 7. | |
| 孙伟博, 王 璞, 杨梓堃, 等. 转Bt基因南林895杨对美国白蛾抗虫性研究. 中南林业科技大学学报, 2020, 40 (7): 107- 118. | |
| Sun W B, Wang P, Yang Z K, et al. Study on the insect resistance of Bt transgenic poplar Nanlin 895 to Hyphantria cunea. Journal of Central South University of Forestry & Technology, 2020, 40 (7): 107- 118. | |
| 魏瑞研, 张卫华, 徐 放, 等. 红锥生长性状的全基因组选择与优良子代早期评选. 林业科学, 2024, 60 (12): 83- 91. | |
| Wei R Y, Zhang W H, Xu F, et al. Genomic selection for growth traits and early selection of superior progeny in Castanopsis hystrix. Scientia Silvae Sinicae, 2024, 60 (12): 83- 91. | |
| 张苗苗, 王军辉, 卢 楠, 等. 林木全基因组选择研究现状和应用. 世界林业研究, 2021, 34 (4): 26- 32. | |
| Zhang M M, Wang J H, Lu N, et al. Research progress and application of whole genome selection in forest tree breeding. World Forestry Research, 2021, 34 (4): 26- 32. | |
| 朱 嵊, 黄敏仁. 基因组选择在林木遗传育种研究中的进展与展望. 林业科学, 2020, 56 (11): 176- 186. | |
| Zhu S, Huang M R. Recent advances and prospect of the genomic selection in forest genetics and tree breeding. Scientia Silvae Sinicae, 2020, 56 (11): 176- 186. | |
|
Alemu A, Åstrand J, Montesinos-López O A, et al. Genomic selection in plant breeding: key factors shaping two decades of progress. Molecular Plant, 2024, 17 (4): 552- 578.
doi: 10.1016/j.molp.2024.03.007 |
|
|
Alves F C, Balmant K M, Resende M F R, et al. Accelerating forest tree breeding by integrating genomic selection and greenhouse phenotyping. The Plant Genome, 2020, 13 (3): e20048.
doi: 10.1002/tpg2.20048 |
|
| Boopathi N M. 2020. Genetic mapping and marker assisted selection. Singapore: Springer. | |
|
Crossa J, Pérez-Rodríguez P, Cuevas J, et al. Genomic selection in plant breeding: methods, models, and perspectives. Trends in Plant Science, 2017, 22 (11): 961- 975.
doi: 10.1016/j.tplants.2017.08.011 |
|
|
Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics, 2011, 27 (15): 2156- 2158.
doi: 10.1093/bioinformatics/btr330 |
|
|
Desta Z A, Ortiz R. Genomic selection: genome-wide prediction in plant improvement. Trends in Plant Science, 2014, 19 (9): 592- 601.
doi: 10.1016/j.tplants.2014.05.006 |
|
|
Duarte D, Jurcic E J, Dutour J, et al. Genomic selection in forest trees comes to life: unraveling its potential in an advanced four-generation Eucalyptus grandis population. Frontiers in Plant Science, 2024, 15, 1462285.
doi: 10.3389/fpls.2024.1462285 |
|
| Grattapaglia D, Resende M D V. Genomic selection in forest tree breeding. Tree Genetics & Genomes, 2011, 7 (2): 241- 255. | |
|
Grattapaglia D. Twelve years into genomic selection in forest trees: climbing the slope of enlightenment of marker assisted tree breeding. Forests, 2022, 13 (10): 1554.
doi: 10.3390/f13101554 |
|
|
Habier D, Fernando R L, Dekkers J C M. The impact of genetic relationship information on genome-assisted breeding values. Genetics, 2007, 177 (4): 2389- 2397.
doi: 10.1534/genetics.107.081190 |
|
|
Habier D, Fernando R L, Kizilkaya K, et al. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics, 2011, 12 (1): 186.
doi: 10.1186/1471-2105-12-186 |
|
|
Li Z T, Sillanpää M J. Dynamic quantitative trait locus analysis of plant phenomic data. Trends in Plant Science, 2015, 20 (12): 822- 833.
doi: 10.1016/j.tplants.2015.08.012 |
|
| McLean D, Apiolaza L, Paget M, et al. Simulating deployment of genetic gain in a radiata pine breeding program with genomic selection. Tree Genetics & Genomes, 2023, 19 (4): 33. | |
|
Meuwissen T H E, Hayes B J, Goddard M E. Prediction of total genetic value using genome-wide dense marker maps. Genetics, 2001, 157 (4): 1819- 1829.
doi: 10.1093/genetics/157.4.1819 |
|
| Muranty H, Jorge V, Bastien C, et al. Potential for marker-assisted selection for forest tree breeding: lessons from 20 years of MAS in crops. Tree Genetics & Genomes, 2014, 10 (6): 1491- 1510. | |
| Porth I, Klápště J, McKown A. 2024. The poplar genome. Cham: Springer International Publishing. | |
|
Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics, 2007, 81 (3): 559- 575.
doi: 10.1086/519795 |
|
|
Simiqueli G F, Resende R T, Takahashi E K, et al. Realized genomic selection across generations in a reciprocal recurrent selection breeding program of Eucalyptus hybrids. Frontiers in Plant Science, 2023, 14, 1252504.
doi: 10.3389/fpls.2023.1252504 |
|
|
Sun M Y, Zhang M Y, Kumar S, et al. Genomic selection of eight fruit traits in pear. Horticultural Plant Journal, 2024, 10 (2): 318- 326.
doi: 10.1016/j.hpj.2023.04.008 |
|
|
VanRaden P M. Efficient methods to compute genomic predictions. Journal of Dairy Science, 2008, 91 (11): 4414- 4423.
doi: 10.3168/jds.2007-0980 |
|
| Varshney R K, Roorkiwal M, Sorrells M E. 2017. Genomic selection for crop improvement. Cham: Springer International Publishing. | |
|
Wang Q, Jiang S, Li T, et al. G2P provides an integrative environment for multi-model genomic selection analysis to improve genotype-to-phenotype prediction. Frontiers in Plant Science, 2023, 14, 1207139.
doi: 10.3389/fpls.2023.1207139 |
|
|
Wei S Y, Yang G, Yang Y H, et al. Time-sequential detection of quantitative trait loci and candidate genes underlying the dynamic growth of Salix suchowensis. Tree Physiology, 2022, 42 (4): 877- 890.
doi: 10.1093/treephys/tpab138 |
|
|
Wickham H. ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics, 2011, 3 (2): 180- 185.
doi: 10.1002/wics.147 |
|
|
Wu R L, Ma C X, Yang M C K, et al. Quantitative trait loci for growth trajectories in Populus. Genetical Research, 2003, 81 (1): 51- 64.
doi: 10.1017/S0016672302005980 |
|
|
Xia H, Hao Z Y, Shen Y F, et al. Genome-wide association study of multiyear dynamic growth traits in hybrid Liriodendron identifies robust genetic loci associated with growth trajectories. The Plant Journal, 2023, 115 (6): 1544- 1563.
doi: 10.1111/tpj.16337 |
|
|
Xue L J, Wu H T, Chen Y N, et al. Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nature Communications, 2020, 11 (1): 5893.
doi: 10.1038/s41467-020-19559-2 |
|
|
Yang J, Lee S H, Goddard M E, et al. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics, 2011, 88 (1): 76- 82.
doi: 10.1016/j.ajhg.2010.11.011 |
|
| Yin L L, Zhang H H, Tang Z S, et al. 2021. rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics, Proteomics & Bioinformatics, 19(4): 619–628. |
| [1] | Junhui Wang,Changjun Ding,Wei Li,Keming Luo,Jun Wang,Weixi Zhang,Shihui Niu,Miaomiao Zhang,Xiyang Zhao,Liangjiao Xue,Hengfu Yin. Advances in Genetic Breeding Research of Chinese Forest Trees in 2024 [J]. Scientia Silvae Sinicae, 2025, 61(7): 35-51. |
| [2] | Lei Zhang,Xinglu Zhou,Lijuan Wang,Jianjun Hu. Advance of Poplar Molecular Breeding with Insect Resistance and Transgenic Biosafety Assessment Research [J]. Scientia Silvae Sinicae, 2025, 61(2): 190-203. |
| [3] | Qian Li,Fan Zhang,Xiangxue Meng,Xiaoyun Wu,Jianzhuang Pang,Hang Xu,Zhiqiang Zhang. Simulation of Net Carbon Exchange of Poplar Plantation in North China Plain Based on Decomposition-Reconstruction and Machine Learning [J]. Scientia Silvae Sinicae, 2025, 61(12): 72-82. |
| [4] | Jiangtao Guo,Jingjing Peng,Xin Ma,Fan Liu,Yongxiu Xia. Responses of Poplar Plantations in Northern China Sandy Land to Conventional Fertilization and Drip Fertigation [J]. Scientia Silvae Sinicae, 2025, 61(10): 135-145. |
| [5] | Lingyu Yang,Wenguang Shi,Zhibin Luo. Characteristics of Ectomycorrhizal Fungi Paxillus involutus Promoting Nitrogen Uptake and Utilization of Its Host Populus tremula × Populus alba [J]. Scientia Silvae Sinicae, 2024, 60(9): 69-79. |
| [6] | Xiaolin Qiu,Shumin Wang,Lu Yu,Yuchen Yang,Dianguang Xiong,Chengming Tian. Functional of SNARE Protein CcNyv1 in Cytospora chrysosperma [J]. Scientia Silvae Sinicae, 2024, 60(9): 90-98. |
| [7] | Yingqi He,Lufei Wang,Yamei Zhang,Yanglun Yu,Wenji Yu. Effect of Compression Ratios on the Surface Hardness of Poplar Wood Scrimber [J]. Scientia Silvae Sinicae, 2024, 60(9): 141-149. |
| [8] | Aoyu Wang,Youzheng Guo,Tan Deng,Yang Liu,Nan Di,Jie Duan,Ximeng Li,Benye Xi. Comparison of Several Methods for Evaluating Plant Water Regulation Strategies [J]. Scientia Silvae Sinicae, 2024, 60(8): 109-119. |
| [9] | Qing Zhou,Heng Zhang,Pengwu Zhao,Yong Zhou,Lin Zhang,Hongzhuo Mi,Jiafu Wang,Mengyu Zhao,Zehua Yang. Differences in the Orobability and Drivers of Forest Fires in the Daxing’an Mountains of Inner Mongolia before and after the Major Historical Event of the Forest Fire in 1987 [J]. Scientia Silvae Sinicae, 2024, 60(7): 81-94. |
| [10] | Kong Yue,Xiang Li,Xinlei Shi,Xuekai Jiao,Peng Wu,Zhongfeng Zhang,Guoliang Dong,Yuanjin Fang. Effects of Thermal Pretreatment on Lateral Performance of Poplar Cross-Laminated Timber Shear Walls [J]. Scientia Silvae Sinicae, 2024, 60(7): 117-128. |
| [11] | Lei Xu,Xiaoyun Wu,Jiang Lü,Yun Shi,Mengxun Zhu,Hang Xu,Zhiqiang Zhang. Impacts of Diffuse Radiation Fraction on Energy Partitioning in a Poplar Plantation in the North China Plain [J]. Scientia Silvae Sinicae, 2024, 60(3): 100-110. |
| [12] | Ruiyan Wei,Weihua Zhang,Fang Xu,Yuanzhen Lin. Genomic Selection for Growth Traits and Early Selection of Superior Progeny in Castanopsis hystrix [J]. Scientia Silvae Sinicae, 2024, 60(12): 83-91. |
| [13] | Meihong Liu,Qiming Yan,Longbo Zi,Yafang Lei,Li Yan. Physical and Mechanical Properties of Furfuryl Alcohol-Epoxidized Vegetable Oils Composite Modified Poplar Wood [J]. Scientia Silvae Sinicae, 2024, 60(11): 149-159. |
| [14] | Jiaming Wan,Jiang Lü,Yun Shi,Hang Xu,Zhiqiang Zhang. Effects of Diffuse Radiation on the Gross Primary Productivity of a Poplar Plantation [J]. Scientia Silvae Sinicae, 2023, 59(5): 1-10. |
| [15] | Lu Han,Han Zhao,Wei Wang,Wenhui Liu,Zaimin Jiang,Jing Cai. Hydraulic Vulnerability Segmentation and Its Correlation with Growth in Hybrid Poplar [J]. Scientia Silvae Sinicae, 2023, 59(3): 94-103. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||