

Scientia Silvae Sinicae ›› 2025, Vol. 61 ›› Issue (10): 111-120.doi: 10.11707/j.1001-7488.LYKX20250316
• Research papers • Previous Articles
Chengshuo Song1,2,Kean-Jin Lim1,2,Erpei Lin1,2,Huahong Huang1,2,*( )
)
Received:2025-05-18
Online:2025-10-25
Published:2025-11-05
Contact:
Huahong Huang
E-mail:huanghh@zafu.edu.cn
CLC Number:
Chengshuo Song,Kean-Jin Lim,Erpei Lin,Huahong Huang. Molecular Identification of ClBFN Gene Family and Anatomical Characteristics of the Sapwood-Heartwood Transition Zone in Cunninghamia lanceolata[J]. Scientia Silvae Sinicae, 2025, 61(10): 111-120.
Table 1
Primers used for qRT-PCR"
| 引物名称 Primer name | 序列(5'–3') Sequence (5'–3') |
| ClEF1α-F | TGGCAAGGAGCTTGAGAAAGAACCCA |
| ClEF1α-R | ACCCCAACAGCAACAGTCTGACGCAT |
| ClBFN1-F | GTGACACCGATGACCATGCT |
| ClBFN1-R | GCGATTGAGTGTTGCAGCCA |
| ClBFN2-F | GATCTGGCTTCTGTCTGCTCAT |
| ClBFN2-R | CAACGCACATGTCTTCTTCTCC |
| ClBFN3-F | TGCCCAAATCCGTATGCAGT |
| ClBFN3-R | ACCTCACAGCTCCTTTTGCA |
| ClBFN4-F | AAAGAGACGTGTGTGTGGCA |
| ClBFN4-R | TCCTCCTGCATCTCCAGTGA |
Table 2
SqRT-PCR primers for screening the transition zone"
| 引物名称 Primer name | 序列(5'–3') Sequence (5'–3') |
| ClEF1α-F | TGGCAAGGAGCTTGAGAAAGAACCCA |
| ClEF1α-R | ACCCCAACAGCAACAGTCTGACGCAT |
| ClBFN1-SF | TTGTTCATGGGCGGATCACA |
| ClBFN1-SR | AGGACAAGCATGGTCATCGG |
| ClBFN3-SF | GCATCTGTTTGTTCCTGGGC |
| ClBFN3-SR | ACTGCATACGGATTTGGGCA |
| ClBFN4-SF | TGCACTCCACATCTCAACCC |
| ClBFN4-SR | TCCTCCTGCATCTCCAGTGA |
Table 3
Physicochemical characteristics of ClBFN proteins in Chinese fir"
| 蛋白质 Protein | 氨基酸残基数 Length/aa | 分子量 Molecular weight /kDa | 等电点 pI | 不稳定系数 Instability index | 总平均疏水性 GRAVY | 亚细胞定位 Subcellular localization |
| ClBFN1 | 305 | 34.88 | 5.43 | 39.82 | ?0.399 | Nucleus |
| ClBFN2 | 303 | 33.88 | 4.96 | 42.25 | ?0.155 | Cell wall |
| ClBFN3 | 287 | 31.95 | 5.40 | 32.95 | ?0.080 | Cell wall |
| ClBFN4 | 302 | 34.06 | 6.85 | 36.26 | ?0.123 | Cell wall |

Fig.4
Expression profile of ClBFN genes A: Plant materials used for the qPCR analysis;B: Relative expression levels of ClBFN genes in different tissues (the root is used as a reference, and error bar represents standard deviation of three replicates). N: Young needle;ON: Mature needle;X: Xylem;B: Bark;R: Root."
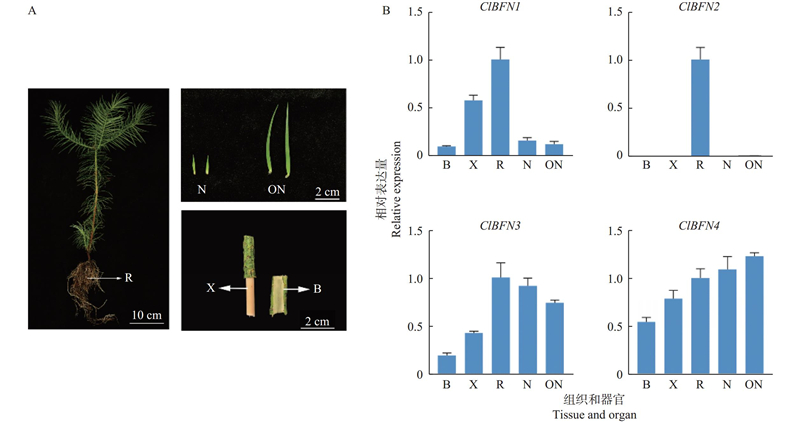

Fig.7
SqRT-PCR and qRT-PCR analysis of ClBFN1 in Tree 1, Tree 2 and Tree 3 SqRT-PCR analysis of ClBFN1(Col. A), CIEF1α((Col. B) in different annual rings; qRT-PCR analysis of CIBFN1(Col. C) in different annual rings. SW is used as a reference. M: DL2000-DNAmarker; 5–8: Different annual ring samples in the direction from the dry-wet demarcation line of the wood core towards the bark, respectively; TZ: Transition zone; SW: Sapwood; ** indicate significant difference at 0. 01 level."
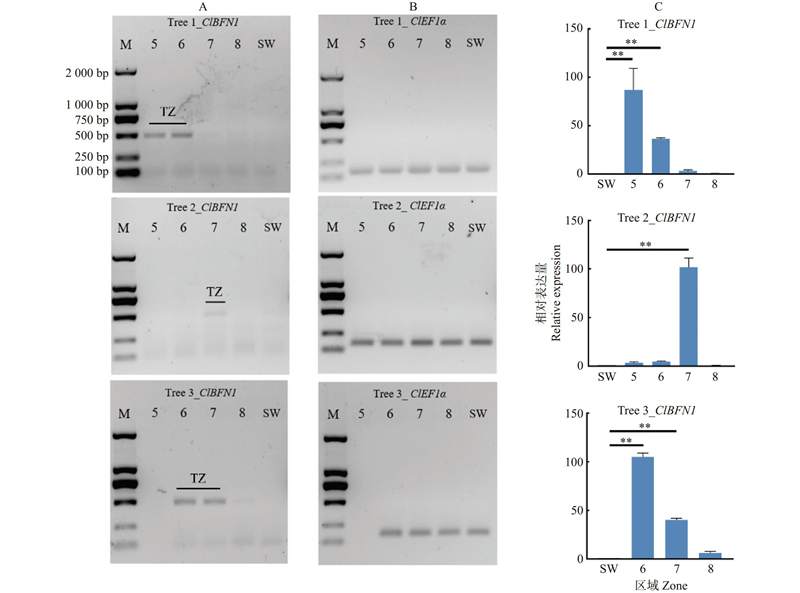

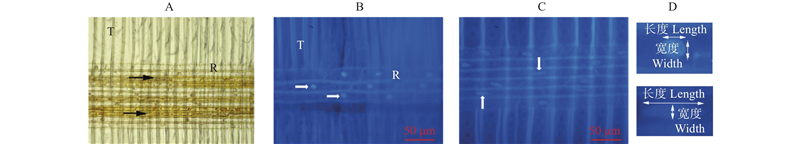
Fig.8
Changes of nuclear characteristics in ray parenchyma cells A: Heartwood in brightfield;B: Transition zone stained by DAPI;C: Sapwood stained by DAPI;D: Schematic diagram of cell nuclear measurement;T: Tracheid;R: Ray parenchyma cell;Black arrows indicate filled ray parenchyma cells, and white arrows point to nucleus nuclei;Bar = 50 μm."
| 常建国, 李新平, 刘世荣, 等. 2009. 油松心边材量及年轮数的变异特征. 林业科学, 45(11): 76–82. | |
| Chang J G, Li X P, Liu S R, et al. 2009. Variations in amount and ring number of sapwood and heartwood of Pinus tabulaeformis. Scientia Silvae Sinicae, 45(11): 76–82.[in Chinese ] | |
| 刘家霖, 王传宽, 张全智. 2014. 不同分化等级兴安落叶松树干心材和边材的空间变异. 林业科学, 50(12): 114–121. | |
| Liu J L, Wang C K, Zhang Q Z. 2014. Spatial variations in stem heartwood and sapwood for Larix gmelinii trees with various differentiation classes. Scientia Silvae Sinicae, 50(12): 114–121. [in Chinese] | |
| 魏明科, 俞金健, 黄晓龙, 等. 2018. 杉木NAC转录因子基因ClNAC1的克隆、表达及单核苷酸多态性分析. 林业科学, 54(9): 49–59. | |
| Wei M K, Yu J J, Huang X L, et al. 2018. Cloning, expression and single nucleotide polymorphisms analysis of NAC transcription factor gene ClNAC1 in Cunninghamia lanceolata. Scientia Silvae Sinicae, 54(9): 49–59. [in Chinese] | |
|
Arakawa I, Funada R, Nakaba S. Changes in the morphology and functions of vacuoles during the death of ray parenchyma cells in Cryptomeria japonica. Journal of Wood Science, 2018, 64 (3): 177- 185.
doi: 10.1007/s10086-017-1692-6 |
|
|
Bergström B. Chemical and structural changes during heartwood formation in Pinus sylvestris. Forestry: An International Journal of Forest Research, 2003, 76 (1): 45- 53.
doi: 10.1093/forestry/76.1.45 |
|
|
Beritognolo I, Magel E, Abdel-Latif A, et al. Expression of genes encoding chalcone synthase, flavanone 3-hydroxylase and dihydroflavonol 4-reductase correlates with flavanol accumulation during heartwood formation in Juglans nigra. Tree Physiology, 2002, 22 (5): 291- 300.
doi: 10.1093/treephys/22.5.291 |
|
|
Cao S, Deng H Y, Zhao Y, et al. Metabolite profiling and transcriptome analysis unveil the mechanisms of red-heart Chinese fir [Cunninghamia lanceolata (Lamb.) Hook] heartwood coloration. Frontiers in Plant Science, 2022, 13, 854716.
doi: 10.3389/fpls.2022.854716 |
|
|
Celedon J M, Bohlmann J. An extended model of heartwood secondary metabolism informed by functional genomics. Tree Physiology, 2018, 38 (3): 311- 319.
doi: 10.1093/treephys/tpx070 |
|
|
Chang S J, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter, 1993, 11 (2): 113- 116.
doi: 10.1007/BF02670468 |
|
|
Escamez S, Tuominen H. Programmes of cell death and autolysis in tracheary elements: when a suicidal cell arranges its own corpse removal. Journal of Experimental Botany, 2014, 65 (5): 1313- 1321.
doi: 10.1093/jxb/eru057 |
|
|
Farage-Barhom S, Burd S, Sonego L, et al. Localization of the Arabidopsis senescence- and cell death-associated BFN1 nuclease: from the er to fragmented nuclei. Molecular Plant, 2011, 4 (6): 1062- 1073.
doi: 10.1093/mp/ssr045 |
|
|
Fendrych M, Van Hautegem T, Van Durme M, et al. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Current Biology, 2014, 24 (9): 931- 940.
doi: 10.1016/j.cub.2014.03.025 |
|
|
Fukuda H. Xylogenesis: initiation, progression, and cell death. Annual Review of Plant Physiology and Plant Molecular Biology, 1996, 47, 299- 325.
doi: 10.1146/annurev.arplant.47.1.299 |
|
| Fukuda H. Tracheary element differentiation. Plant Cell, 1997, 9 (7): 1147- 1156. | |
|
Gunawardena A H L A N. Programmed cell death and tissue remodelling in plants. Journal of Experimental Botany, 2008, 59 (3): 445- 451.
doi: 10.1093/jxb/erm189 |
|
|
Han X J, Zhou Y F, Ni X L, et al. Programmed cell death during the formation of rhytidome and interxylary cork in roots of Astragalus membranaceus (Leguminosae). Microscopy Research and Technique, 2021, 84 (7): 1400- 1413.
doi: 10.1002/jemt.23696 |
|
|
Huang H H, Xu L L, Tong Z K, et al. De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genomics, 2012, 13, 648.
doi: 10.1186/1471-2164-13-648 |
|
| Kampe A, Magel E. 2013. New insights into heartwood and heartwood formation// Fromm J. Cellular aspects of wood formation. Berlin, Heidelberg: Springer Berlin Heidelberg, 71–95. | |
|
Kim M H, Bae E K, Lee H, et al. Current understanding of the genetics and molecular mechanisms regulating wood formation in plants. Genes, 2022, 13 (7): 1181.
doi: 10.3390/genes13071181 |
|
|
Krela R, Poreba E, Lesniewicz K. Variations in the enzymatic activity of S1-type nucleases results from differences in their active site structures. Biochimica et Biophysica Acta (BBA)–General Subjects, 2023, 1867 (10): 130424.
doi: 10.1016/j.bbagen.2023.130424 |
|
|
Lesniewicz K, Karlowski W M, Pienkowska J R, et al. The plant s1-like nuclease family has evolved a highly diverse range of catalytic capabilities. Plant Cell Physiology, 2013, 54 (7): 1064- 1078.
doi: 10.1093/pcp/pct061 |
|
|
Lim K J, Paasela T, Harju A, et al. Developmental changes in Scots pine transcriptome during heartwood formation. Plant Physiology, 2016, 172 (3): 1403- 1417.
doi: 10.1104/pp.16.01082 |
|
| Luigi D F, Magel E. Identification of biochemical differences between the sapwood and transition zone in Robinia pseudoacacia L. by differential display of proteins. Holzforschung, 2012, 66 (4): 543- 549. | |
| Magel E. 2000. Biochemistry and physiology of heart wood formation// Savidge R, Barnett J, Napier R. Molecular and Cell Biology of BIOS. Oxford: Scientific Publishers. | |
|
Moshchenskaya Y L, Galibina N A, Nikerova K M, et al. Plant-programmed cell death-associated genes participation in Pinus sylvestris L. trunk tissue formation. Plants, 2022, 11 (24): 3438.
doi: 10.3390/plants11243438 |
|
|
Nakaba S, Kubo T, Funada R. Differences in patterns of cell death between ray parenchyma cells and ray tracheids in the conifers Pinus densiflora and Pinus rigida. Trees, 2008, 22 (5): 623- 630.
doi: 10.1007/s00468-008-0220-0 |
|
|
Nakaba S, Kubo T, Funada R. Nuclear DNA fragmentation during cell death of short-lived ray tracheids in the conifer Pinus densiflora. Journal of Plant Research, 2011, 124 (3): 379- 384.
doi: 10.1007/s10265-010-0384-8 |
|
|
Nakaba S, Sano Y, Kubo T, et al. The positional distribution of cell death of ray parenchyma in a conifer, Abies sachalinensis. Plant Cell Reports, 2006, 25 (11): 1143- 1148.
doi: 10.1007/s00299-006-0194-6 |
|
|
Niu S H, Li J, Bo W H, et al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell, 2022, 185 (1): 204- 217.
doi: 10.1016/j.cell.2021.12.006 |
|
|
Perez-Amador M A, Abler M L, De Rocher E J, et al. Identification of BFN1, a bifunctional nuclease induced during leaf and stem senescence in Arabidopsis. Plant Physiology, 2000, 122 (1): 169- 180.
doi: 10.1104/pp.122.1.169 |
|
|
Plomion C, Leprovost G, Stokes A. Wood formation in trees. Plant Physiology, 2001, 127 (4): 1513- 1523.
doi: 10.1104/pp.010816 |
|
|
Son N T. A review on the medicinal plant Dalbergia odorifera species: phytochemistry and biological activity. Evidence-Based Complementary and Alternative Medicine, 2017, 2017, 7142370.
doi: 10.1155/2017/7142370 |
|
| Song K L, Liu B, Jiang X M, et al. Cellular changes of tracheids and ray parenchyma cells from cambium to heartwood in Cunninghamia lanceolata. Journal of Tropical Forest Science, 2011, 23 (4): 478- 487. | |
| Spicer R. 2005. Senescence in secondary xylem: heartwood formation as an active developmental program//Holbrook N M, Zwieniecki M A. Vascular transport in plants. Burlington: Academic Press, 457–475. | |
|
Sugiyama M, Ito J, Aoyagi S, et al. Endonucleases. Plant Molecular Biology, 2000, 44 (3): 387- 397.
doi: 10.1023/A:1026504911786 |
|
|
Yang X Q, Yu X R, Liu Y, et al. Comparative metabolomics analysis reveals the color variation between heartwood and sapwood of Chinese fir (Cunninghamia lanceolata (Lamb.) Hook. Industrial Crops and Products, 2021, 169, 113656.
doi: 10.1016/j.indcrop.2021.113656 |
|
|
Yeh T F, Chu J H, Liu L Y, et al. Differential gene profiling of the heartwood formation process in Taiwania cryptomerioides Hayata xylem tissues. International Journal of Molecular Sciences, 2020, 21 (3): 960.
doi: 10.3390/ijms21030960 |
|
| Zhang X, Huang K, Ye Y J, et al. 2015. Biomedical molecular of woody extractives of Cunninghamia lanceolata biomass. Pakistan Journal of Pharmaceutical Sciences, 28(2 Suppl): 761–764. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||