

Scientia Silvae Sinicae ›› 2025, Vol. 61 ›› Issue (2): 131-141.doi: 10.11707/j.1001-7488.LYKX20240119
• Research papers • Previous Articles Next Articles
Mingtong Liu,Hebi Zhuang,Zitong Wang,Shuaibin Shi,Xiaojuan Liu,Erpei Lin,Xiange Hu,Huahong Huang*( )
)
Received:2024-03-01
Online:2025-02-25
Published:2025-03-03
Contact:
Huahong Huang
E-mail:huanghh1976@163.com
CLC Number:
Mingtong Liu,Hebi Zhuang,Zitong Wang,Shuaibin Shi,Xiaojuan Liu,Erpei Lin,Xiange Hu,Huahong Huang. Biological Functional Analysis of Transcription Factor Gene ClNAC40 Regulating Secondary Cell Wall Development in Cunninghamia lanceolata[J]. Scientia Silvae Sinicae, 2025, 61(2): 131-141.
Table 2
Primers used for RT-qPCR"
| 引物名称 Primer name | 序列 Sequence (5'→3') | 引物名称 Primer name | 序列 Sequence (5'→3') | |
| ClNAC40-QF | TATCAGACAGTGGTAAAGGCT | AtCAD5-QF | TTGGCTGATTCGTTGGATTA | |
| ClNAC40-QR | TATCAGACAGTGGTAAAGGCT | AtCAD5-QR | ATCACTTTCCTCCCAAGCAT | |
| ClActin1-QF | GAGGGACCAGATTCATCGTATTC | AtCOMT1-QF | CTGCTACGAGTCACTTCCAGAGG | |
| ClActin1-QR | ATGCTGGTATTGCTGATCGTATG | AtCOMT1-QR | GAGTAACTCAATAAGGTTAACACC | |
| AtActin2-QF | CCTGAAAGGAAGTACAGTG | AtHCT-QF | GCCTGCACCAAGTATGAAGA | |
| AtActin2-QR | CTGTGAACGATTCCTGGAC | AtHCT-QR | GACAGTGTTCCCATCCTCCT | |
| AtPAL1-QF | AAGATTGGAGCTTTCGAGGA | AtC3H1-QF | GTTGGACTTGACCGGATCTT | |
| AtPAL1-QR | TCTGTTCCAAGCTCTTCCCT | AtC3H1-QR | ATTAGAGGCGTTGGAGGATG | |
| AtPAL2-QF | GAGGCAGCGTTAAGGTTGAG | AtCCoAOMT1-QF | CTCAGGGAAGTGACAGCAAA | |
| AtPAL2-QR | TTCTCGGTTAGCGATTCACC | AtCCoAOMT1-QR | GTGGCGAGAAGAGAGTAGCC | |
| AtC4H-QF | ACTGGCTTCAAGTCGGAGAT | AtCCR1-QF | GTGCAAAGCAGATCTTCAGG | |
| AtC4H-QR | ACACGACGTTTCTCGTTCTG | AtCCR1-QR | GCCGCAGCATTAATTACAAA | |
| At4CL1-QF | TCAACCCGGTGAGATTTGTA TCGTCATCGATCAATCCAAT | AtF5H-QF | CTTCAACGTAGCGGATTTCA | |
| At4CL1-QR | TCGTCATCGATCAATCCAAT | AtF5H-QR | AGATCATTACGGGCCTTCAC |
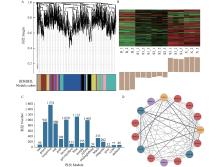
Fig.1
Gene co-expression network A: Gene cluster tree, each cluster tree corresponds to a gene module, and the modules with the same clustering level are represented by the same color. B: Gene expression profiles of tan module. C: The 15 modules clustered and the number of genes in each module. D: Gene co-expression network of tan module. B (at the horizontal axis in Figure 1B): Bark; S1–S3: Stem in different maturities; X: Xylem; The red node represents transcription factor, the blue node represents genes related to lignin synthesis, the orange node represents genes related to cellulose synthesis and the purple node represents genes related to hemicellulose synthesis; The thickness of the line represents the magnitude of the correlation value."


Fig.5
Analysis of ClNAC40 genes expression in different tissues and during compression wood formation A: Relative expression analysis of ClNAC40 in different tissues, the S1 is used as a reference; B: Relative expression analysis of ClNAC40 in compression wood, the CK is used as a reference. Error bar represents standard deviation of three replicates. The same small letter above columns indicates no significant difference at 0.05 levels. RT: Root; FC: Female cone; MC: Male cone; L: Leave; S1–S3: Stem in different maturities; XY: Xylem; BK: Bark; CW10、CW30、CW60: Compression wood with oblique treatment for 10, 30, and 60 days; CK: Upright wood; OW10、OW30、OW60: Opposite wood with oblique treatment for 10, 30, and 60 days."
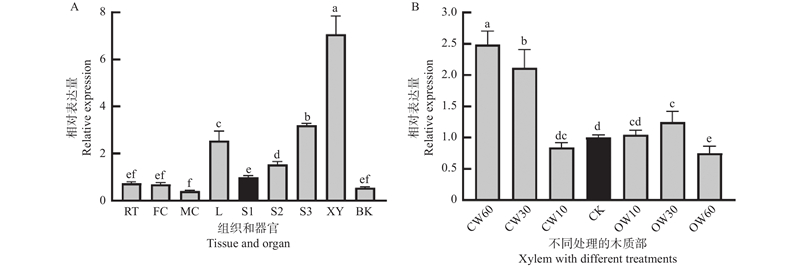
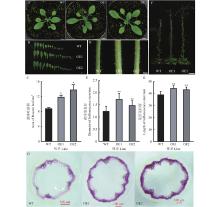
Fig.6
Overexpression of ClNAC40 phenotype in Arabidopsis thaliana A: Phenotypes of 4-week-old wild-type (WT) and overexpressing ClNAC40 Arabidopsis plants (OE1, OE2); B, C: Number and area of rosette leaves in 4-week-old wild-type (WT) and overexpressing ClNAC40 Arabidopsis plants (OE1, OE2); D, E: Inflorescence stem diameter of 4-week-old wild-type (WT) and overexpressing ClNAC40 Arabidopsis plants (OE1, OE2); F, G: Phenotype and inflorescence stem height of 8-week-old wild-type (WT) and overexpressing ClNAC40 Arabidopsis plants (OE1, OE2); H: Inflorescence stem sections of wild-type (WT) and overexpressing ClNAC40 Arabidopsis plants (OE1, OE2); * indicates significant difference compared to WT (P<0.05); ** indicate extremely significant difference compared to WT (P<0.01)."


Fig.7
The relative expression analysis of lignin biosynthesis genes. WT: Wild-type, served as a reference; OE1, OE2: Overexpressing ClNAC40 Arabidopsis plants; * indicates significant difference between OE1 or OE2 and WT (P<0.05); ** indicate extremely significant difference between OE1 or OE2 and WT (P<0.01)."

| 陈亚娟, 王宏芝, 李瑞芬, 等. 毛白杨纤维素合酶基因家族部分成员的克隆及表达. 林业科学, 2011, 47 (10): 70- 75. | |
| Chen Y J, Wang H Z, Li R F, et al. Isolation and expression profile of some members of cellulose synthase gene family in Populus tomentosa. Scientia Silvae Sinicae, 2011, 47 (10): 70- 75. | |
| 程健弘, 魏明科, 林二培, 等. 杉木HD-Zip Ⅲ 转录因子的克隆及表达分析. 农业生物技术学报, 2017, 25 (11): 1820- 1830. | |
| Cheng J H, Wei M K, Lin E P, et al. Cloning and expression analysis of HD-Zip Ш transcriptional factors in Cunninghamia lanceolata. Journal of Agricultural Biotechnology, 2017, 25 (11): 1820- 1830. | |
| 杜明秋, 钟珊丽, 林二培, 等. 2022. 杉木应压木形成中的显微特征及主要代谢成分变化. 核农学报, 36(11): 2307–2315. | |
| Du M Q, Zhong S L, Lin E P, et al. 2022. Anatomy characteristics and study of alterations of key metabolic components in Cunninghamia lanceolata during compression wood formation. Journal of Nuclear Agricultural Sciences, 36(11): 2307–2315. [in Chinese] | |
| 高文杰, 刘 娇, 马祥庆, 等. 杉木NAC基因家族基因的鉴定及生物信息学分析. 中南林业科技大学学报, 2022, 42 (2): 108- 118. | |
| Gao W J, Liu J, Ma X Q, et al. Identification and bioinformatics analysis of Chinese fir NAC gene family. Journal of Central South University of Forestry & Technology, 2022, 42 (2): 108- 118. | |
| 李 媛, 陈金焕, 金 曌, 等. 毛果杨NAC128基因在次生壁形成中的功能. 林业科学, 2020, 56 (11): 62- 72. | |
| Li Y, Chen J H, Jin Z, et al. Functions of NAC128 gene from Populus trichocarpa in secondary cell wall formation. Scientia Silvae Sinicae, 2020, 56 (11): 62- 72. | |
| 倪 飞, 励文豪, 林二培, 等. 光皮桦MYB基因的克隆及表达和调控分析. 林业科学, 2018, 54 (12): 70- 81. | |
| Ni F, Li W H, Lin E P, et al. Cloning, expression and regulation of MYB genes in Betula luminifera. Scientia Silvae Sinicae, 2018, 54 (12): 70- 81. | |
| 王 钰, 童再康, 黄华宏, 等. 光皮桦总RNA的提取及Actin基因的克隆. 浙江林业科技, 2010, 30 (4): 32- 36. | |
| Wang Y, Tong Z K, Huang H H, et al. Extracting of total RNA from Betula luminifera and the cloning of Actin gene. Journal of Zhejiang Forestry Science and Technology, 2010, 30 (4): 32- 36. | |
| 魏明科, 俞金健, 黄晓龙, 等. 杉木NAC转录因子基因ClNAC1的克隆、表达及单核苷酸多态性分析. 林业科学, 2018, 54 (9): 49- 59. | |
| Wei M K, Yu J J, Huang X L, et al. Cloning, expression and single nucleotide polymorphisms analysis of NAC transcription factor gene ClNAC1 in Cunninghamia lanceolata. Scientia Silvae Sinicae, 2018, 54 (9): 49- 59. | |
| 肖玉菲, 刘海龙, 刘雄盛, 等. 尾叶桉GLU4无性系F5H基因的克隆表达及序列分析. 生物技术, 2018, 28 (3): 205- 211. | |
| Xiao Y F, Liu H L, Liu X S, et al. Cloning, expression and sequence analysis of F5H gene in Eucalyptus urophylla clone GLU4. Biotechnology, 2018, 28 (3): 205- 211. | |
| 谢一青, 李志真, 黄儒珠, 等. 光皮桦基因组DNA提取方法比较. 浙江林学院学报, 2006, 23 (6): 664- 668. | |
| Xie Y Q, Li Z Z, Huang R Z, et al. Comparison of methods of extracting genomic DNA from Betula luminifera. Journal of Zhejiang A & F University, 2006, 23 (6): 664- 668. | |
| 徐莉莉, 童再康, 林二培, 等. 杉木苯丙氨酸解氨酶基因ClPAL的克隆与表达分析. 林业科学, 2013, 49 (12): 64- 72. | |
| Xu L L, Tong Z K, Lin E P, et al. Isolation and expression of CIPAL genes in Chinese fir (Cunninghamia lanceolata). Scientia Silvae Sinicae, 2013, 49 (12): 64- 72. | |
| 俞新妥. 中国杉木90年代的研究进展 Ⅰ. 杉木研究的特点及有关基础研究的综述. 福建林学院学报, 2000, 20 (1): 87- 96. | |
| Yu X T. A summary of the studies on Chinese fir in 1990's I. the distinguishing features of Chinese fir research and research development on basic research. Journal of Forest and Environment, 2000, 20 (1): 87- 96. | |
| Camargo E L, Ployet R, Cassan-Wang H, Mounet F, et al. Digging in wood: new insights in the regulation of wood formation in tree species. Advances in Botanical Research, 2019, 89, 201- 233. | |
|
Chen C J, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13 (8): 1194- 1202.
doi: 10.1016/j.molp.2020.06.009 |
|
|
Chen X H, Wang H T, Li X Y, et al. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4 (4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biology, 2019, 19, 231.
doi: 10.1186/s12870-019-1812-0 |
|
|
Huang H H, Xu L L, Tong Z K, et al. De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genomics, 2012, 13, 648.
doi: 10.1186/1471-2164-13-648 |
|
|
Hussey S G, Mizrachi E, Spokevicius A V, et al. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biology, 2011, 11, 173.
doi: 10.1186/1471-2229-11-173 |
|
|
Mitsuda N, Iwase A, Yamamoto H, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. The Plant Cell, 2007, 19 (1): 270- 280.
doi: 10.1105/tpc.106.047043 |
|
|
Ohtani M, Nishikubo N, Xu B, et al. A NAC domain protein family contributing to the regulation of wood formation in poplar. The Plant Journal, 2011, 67 (3): 499- 512.
doi: 10.1111/j.1365-313X.2011.04614.x |
|
| Sakamoto S, Mitsuda N. 2015. Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant and Cell Physiology, 56(2): 299–310. | |
| Salinas M, Xing S, Höhmann S, et al. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta, 2012, 235 (6): 1171- 1184. | |
|
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 2003, 13 (11): 2498- 2504.
doi: 10.1101/gr.1239303 |
|
|
Singh S, Koyama H, Bhati K K, et al. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. Journal of Plant Research, 2021, 134 (3): 475- 495.
doi: 10.1007/s10265-021-01270-y |
|
|
Sun Y M, Jiang C X, Jiang R Q, et al. A novel NAC transcription factor from Eucalyptus, EgNAC141, positively regulates lignin biosynthesis and increases lignin deposition. Frontiers in Plant Science, 2021, 12, 642090.
doi: 10.3389/fpls.2021.642090 |
|
| Taylor-Teeples M, Lin L, de Lucas M, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature, 2015, 517 (7536): 571- 575. | |
|
Wagner A, Tobimatsu Y, Phillips L, et al. CCoAOMT suppression modifies lignin composition in Pinus radiata. The Plant Journal, 2011, 67 (1): 119- 129.
doi: 10.1111/j.1365-313X.2011.04580.x |
|
| Wang H Z, Dixon R A. 2012. On-off switches for secondary cell wall biosynthesis. Molecular Plant, 5(2): 297–303. | |
|
Wang J F, Wang Y P, Zhang J, et al. The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Horticulture Research, 2021, 8 (1): 214.
doi: 10.1038/s41438-021-00649-1 |
|
| Zhang J, Serra J A, Helariutta Y. 2015. Wood development: growth through knowledge. Nature Plants, 1(5): 15060. | |
|
Zhang J, Xie M, Tuskan G A, et al. Recent advances in the transcriptional regulation of secondary cell wall biosynthesis in the woody plants. Frontiers in Plant Science, 2018, 9, 1535.
doi: 10.3389/fpls.2018.01535 |
|
|
Zhang X, Long Y, Chen X X, et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biology, 2021, 21 (1)
doi: 10.1186/s12870-020-02777-7 |
|
| Zhong R Q, Demura T, Ye Z H. 2006. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. The Plant Cell, 18(11): 3158–3170. | |
|
Zhong R Q, Lee C, Haghighat M, et al. Xylem vessel-specific SND5 and its homologs regulate secondary wall biosynthesis through activating secondary wall NAC binding elements. New Phytologist, 2021, 231 (4): 1496- 1509.
doi: 10.1111/nph.17425 |
|
|
Zhong R Q, Lee C, Zhou J L, et al. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. The Plant Cell, 2008, 20 (10): 2763- 2782.
doi: 10.1105/tpc.108.061325 |
|
|
Zhong R Q, Lee C, Ye Z H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends in Plant Science, 2010, 15 (11): 625- 632.
doi: 10.1016/j.tplants.2010.08.007 |
|
|
Zhong R Q, McCarthy R L, Lee C, et al. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiology, 2011, 157 (3): 1452- 1468.
doi: 10.1104/pp.111.181354 |
|
| Zhong R Q, Ye Z H. 2007. Regulation of cell wall biosynthesis. Current Opinion in Plant Biology, 10: 564–572. | |
| Zhong R Q, Ye Z H. 2015a. Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant and Cell Physiology, 56(2): 195–214. | |
| Zhong R Q, Ye Z H. 2015b. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signaling and Behavior, 10(2): e989746. | |
|
Zhuang H B, Chong S L, Priyanka B, et al. Full-length transcriptomic identification of R2R3-MYB family genes related to secondary cell wall development in Cunninghamia lanceolata (Chinese fir). BMC Plant Biology, 2021, 21 (1): 581.
doi: 10.1186/s12870-021-03322-w |
| [1] | Jiang He,Lin Qin. Climate-Sensitive Tree Recruitment Model for Natural Cunninghamia lanceolata Forests [J]. Scientia Silvae Sinicae, 2025, 61(1): 70-80. |
| [2] | Shuya Yang,Jingru Wang,Yingying Zhu,Lita Yi,Meihua Liu. Effects of Mixed Plantation of Cunninghamia lanceolata and Phoebe chekiangensis on Root Exudates and Community Structure of Arbuscular Mycorrhizal Fungi [J]. Scientia Silvae Sinicae, 2024, 60(9): 59-68. |
| [3] | Linxin Li,Guiyun Yang,Haolan Guo,Qiang Dong,Ming Li,Xiangqing Ma,Pengfei Wu. Effects of Propagation Methods on Biomass, Morphological Traits and Carbon and Nitrogen Contents of Fine Roots at Different Orders of Chinese Fir Seedlings [J]. Scientia Silvae Sinicae, 2024, 60(7): 47-55. |
| [4] | Yingchao Ruan, Rexitahong Subi,Xi Lin,Ming Li,Shaohui Fan,Suiqi Feng,Zhiyun Chen,Xiangqing Ma,Zongming He. Effects of Pruning Intensity on the Formation and Quality of Clear Wood of Trees in Cunninghamia lanceolata Plantations [J]. Scientia Silvae Sinicae, 2024, 60(6): 50-59. |
| [5] | Hui Jia,Min Zhu,Zaipeng Yu,Xiaohua Wan,Yanrong Fu,Sirong Wang,Bingzhang Zou,Zhiqun Huang. Relationship between Litter Production, Litter Turnover Period and Leaf Traits of Different Tree Species in Subtropical Young Afforested Land [J]. Scientia Silvae Sinicae, 2024, 60(1): 12-18. |
| [6] | Wenfei Zhao,Xiaoyu Cao,Zhengchang Xie,Yifan Pang,Yaping Sun,Jiping Li,Yongjun Mo,Da Yuan. Evaluation of Stand Spatial Structure of Cunninghamia lanceolata Public Welfare Forest by Using Structural Equation Model [J]. Scientia Silvae Sinicae, 2022, 58(8): 76-88. |
| [7] | Zhouyang Li,Wenling Lu,Wang Qian,Yizi Huang,Erpei Lin,Huahong Huang,Zaikang Tong. Biological Characteristics and Response to Aluminum Stress of Root Border Cells in Cunninghamia lanceolata and Their Response to Aluminum Stress [J]. Scientia Silvae Sinicae, 2022, 58(7): 73-81. |
| [8] | Sifan Shen,Zhen Zhang,Xiangbo Kong,Fu Liu,Sufang Zhang. Research Techniques of Insect Odour Receptors and Their Application in Forest Insects [J]. Scientia Silvae Sinicae, 2020, 56(5): 150-159. |
| [9] | Xiaoli Yan, Wenjia Hu, Yuanfan Ma, yufan Huo, Tuo Wang, Xiangqing Ma. Nitrogen Uptake Preference of Cunninghamia lanceolata, Pinus massoniana, and Schima superba under Heterogeneous Nitrogen Supply Environment and their Root Foraging Strategies [J]. Scientia Silvae Sinicae, 2020, 56(2): 1-11. |
| [10] | Xia Li,Libao Wang,Yafeng Wen,Jun Lin,Xingtong Wu,Meiling Yuan,Yuan Zhang,Minqiu Wang,Xinyu Li. Genetic Diversity of Chinese Fir (Cunninghamia lanceolata) Breeding Populations among Different Generations [J]. Scientia Silvae Sinicae, 2020, 56(11): 53-61. |
| [11] | Hu Huaying, Zhang Hong, Cao Sheng, Yin Danyang, Zhou Chuifan, He Zongming. Effects of Biochar Application on Soil Bacterial Community Structure and Diversity in Cunninghamia lanceolata Plantations [J]. Scientia Silvae Sinicae, 2019, 55(8): 184-193. |
| [12] | Wu Zhilong, Zhou Chengjun, Zhou Xinnian, Liu Fuwan, Zhu Qixiong, Huang Jinyong, Chen Wen. Difference in Soil Respiration Rates of the Mixed Plantations of Cunninghamia lanceolata and Broadleaved Trees 5 Years after Harvesting at Different Intensities [J]. Scientia Silvae Sinicae, 2019, 55(6): 142-149. |
| [13] | Yin Shuyan, Li Bo, Zhou Chenggang, Zhang Weiguang, Xie Lixia, Liu Yongjie. Analysis on Species Differentiation of Oligonychus ununguis on Castanea mollissima and Cunninghamia lanceolata based on 28S rDNA Gene Sequences [J]. Scientia Silvae Sinicae, 2019, 55(4): 122-128. |
| [14] | Yingkai Zhang,Pengju Liu,Changchun Liu,Yi Ren. Prediction Method of Cunninghamia lanceolata Growth Based on Spatial Clustering [J]. Scientia Silvae Sinicae, 2019, 55(11): 137-144. |
| [15] | Hanbin Wu,Aiguo Duan,Jianguo Zhang. Growth Variation and Selection Effect of Cunninghamia lanceolata Provenances at Different Stand Ages [J]. Scientia Silvae Sinicae, 2019, 55(10): 181-192. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||