

Scientia Silvae Sinicae ›› 2025, Vol. 61 ›› Issue (6): 49-60.doi: 10.11707/j.1001-7488.LYKX20240529
• Research papers • Previous Articles Next Articles
Yue Fei,Junhui Wang,Nan Lu,Miaomiao Zhang*( )
)
Received:2024-09-12
Online:2025-06-10
Published:2025-06-26
Contact:
Miaomiao Zhang
E-mail:mmzhang@caf.ac.cn
CLC Number:
Yue Fei,Junhui Wang,Nan Lu,Miaomiao Zhang. Establishment of a Transient Gene Silencing System in Catalpa bungei Based on PDS Homologous Genes[J]. Scientia Silvae Sinicae, 2025, 61(6): 49-60.
Table 1
Primers for qRT-PCR experiments"
| 引物 Primer | 引物序列(5'–3') Primer sequence (5'–3') | 用途 Purpose |
| qCbuactin_F | TTGTGATGGACTCTGGTGATG | 内参基因Reference gene |
| qCbuactin_R | TGTTCATAGTCAAGAGCCACG | 内参基因Reference gene |
| qCbuPDS-1_F | TGGACAAGGGAGGCAGGT | CbuPDS-1 |
| qCbuPDS-1_R | TTGGACATGGCAAGCCCC | CbuPDS-1 |
| qCbuPDS-2_F | ACAGGCTGGGACCAGGAA | CbuPDS-2 |
| qCbuPDS-2_R | CTCTGTGGCCCAAGCGAA | CbuPDS-2 |
| qCbuPDS-3_F | GCTTTCCACCCACCGGAA | CbuPDS-3 |
| qCbuPDS-3_R | GAGCCTGCGTGACCTGTT | CbuPDS-3 |
| qCbuPDS-4_F | AGCTTTGCACTCAGGCCA | CbuPDS-4 |
| qCbuPDS-4_R | TGGAATGGCTGCCTCTGC | CbuPDS-4 |
| qCbuPDS-5_F | AAGCTCAGGATGGAATAACAG | CbuPDS-5 |
| qCbuPDS-5_R | TTCACACACACAGAGTCTTAC | CbuPDS-5 |
| qCbuPDS-6_F | AAACAGGGTTTTGATGTTACGGTTC | CbuPDS-6 |
| qCbuPDS-6_R | TTTGGAATATTGGATACTCGACCTC | CbuPDS-6 |
Table 3
Species information for sequence alignment and phylogenetic analysis"
| 科 Family | 种 Species | 蛋白名称 Protein name | 登录号 Accession No. |
| 紫葳科Bignoniaceae | 楸Catalpa bungei | CbuPDS-1 | NA |
| CbuPDS-2 | NA | ||
| CbuPDS-3 | NA | ||
| CbuPDS-4 | NA | ||
| CbuPDS-5 | XCI54788 | ||
| CbuPDS-6 | NA | ||
| 紫葳科Bignoniaceae | 紫花风铃木Handroanthus impetiginosus | HiPDS | PIN13733.1 |
| 十字花科Brassicaceae | 拟南芥Arabidopsis thaliana | AtPDS | AEE83394.2 |
| AtPDS-1 | sp|Q07356.1| | ||
| AtPDS-3 | NP_001319934.1 | ||
| AtPDS-4 | AAK64084.1 | ||
| 十字花科Brassicaceae | 羽衣甘蓝Brassica oleracea | BoPDS | ABV46593.1 |
| 番木瓜科Caricaceae | 番木瓜Carica papaya | CpPDS | ABG72807.2 |
| 葫芦科Cucurbitaceae | 墨西哥南瓜Cucurbita argyrosperma | CaPDS | KAG7029123.1 |
| CaPDS-2 | KAG6597678.1 | ||
| 柿科Ebenaceae | 柿Diospyros kaki | DkPDS | ACY78343.1 |
| 杜鹃花科Ericaceae | 羊踯躅Rhododendron molle | RmPDS | APB08594.1 |
| 列当科Orobanchaceae | 地黄Rehmannia glutinosa | RgPDS | QXI69585.1 |
| 罂粟科Papaveraceae | 白屈菜Chelidonium majus | ChmaPDS | UOL49141.1 |
| 蔷薇科Rosaceae | 杏Prunus armeniaca | AprPds | AAX33347.1 |
| 蔷薇科Rosaceae | 草莓Fragaria x ananassa | FaPDS | ACR61393.1 |
| 芸香科Rutaceae | 柚Citrus maxima | CimaPDS | AJT59422.1 |
| 杨柳科Salicaceae | 毛果杨Populus trichocarpa | PtPDS | XP_002321104.4 |
| 茄科Solanaceae | 本氏烟草Nicotiana benthamiana | NbPDS | ABY25272.1 |
| NbPDS-1 | ABE99707.1 | ||
| 茄科Solanaceae | 番茄Solanum lycopersicum | SlPDS | NP_001234095.1 |
| SlPDS-1 | CAB59726.1 | ||
| SlPDS-2 | ABR57230.1 | ||
| 茄科Solanaceae | 矮牵牛Petunia x hybrida | PhPDS | AKM12416.1 |
| 菊科Asteraceae | 万寿菊Tagetes erecta | TePDS | AAM45380.1 |
| 禾本科Gramineae | 小麦Triticum aestivum | TaPDS | ACL36586.1 |
| 禾本科Poaceae | 稻Oryza sativa | OsPDS | AAD02489.1 |
| 禾本科Poaceae | 玉蜀黍Zea mays | ZmPDS | AAA99519.1 |
| 石蒜科Amaryllidaceae | 黄水仙Narcissus pseudonarcissus | NpPDS | CAA55392.1 |
Table 5
Information on CbuPDS genes family members in Catalpa bungei"
| 序号 | 基因号 | 基因名 | 染色体定位 | 氨基酸长度 | 等电点 | 相对分子质量 | 亚细胞定位预测 |
| No. | Gene ID | Gene Name | Chromosomal location | Amino acid length | pI | MW | Subcellular Localization Prediction |
| 1 | evm.model.group16.331 | CbuPDS-1 | Chr 8 | 573 | 8.71 | 63 508.07 | 叶绿体Chloroplast |
| 2 | evm.model.group1.1336 | CbuPDS-2 | Chr 12 | 418 | 5.58 | 46 726.65 | 叶绿体Chloroplast |
| 3 | evm.model.group4.455 | CbuPDS-3 | Chr 15 | 566 | 8.69 | 62 794.17 | 叶绿体Chloroplast |
| 4 | evm.model.group6.920 | CbuPDS-4 | Chr 17 | 286 | 7.6 | 32 457.68 | 细胞核Nucleus |
| 5 | evm.model.group6.921 | CbuPDS-5 | Chr 17 | 315 | 7.01 | 35 187.35 | 叶绿体Chloroplast |
| 6 | evm.model.scaffold247.29 | CbuPDS-6 | NA | 532 | 5.89 | 60 706.42 | 细胞质Cytoplasmcyto |

Fig.8
Construction and application of CbuPDS-5 expression vector A: Schematic representation of the pTRV2-CbuPDS-5 constructs; B: Identification of plasmid DNA from positive clones digested with Xba Ⅰ and Sac Ⅰ;C: Positive test for amplified recombinant vector;D: Abaxial surface of leaves of C. bungei was infected by leaf injection. LB and RB refer to the left and right border sequences of T-DNA. M1 is the DL 15 000 DNA Marker, and M2 is the DL 500 DNA Marker;C1 to C6 refers to Clone 1 to 6."
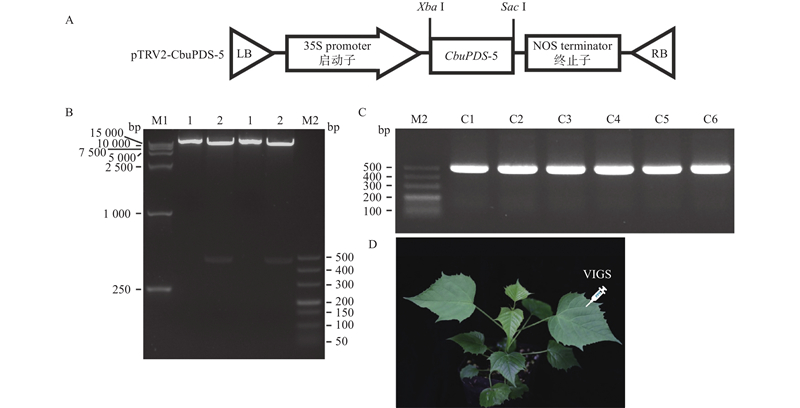
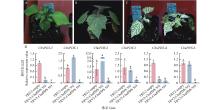
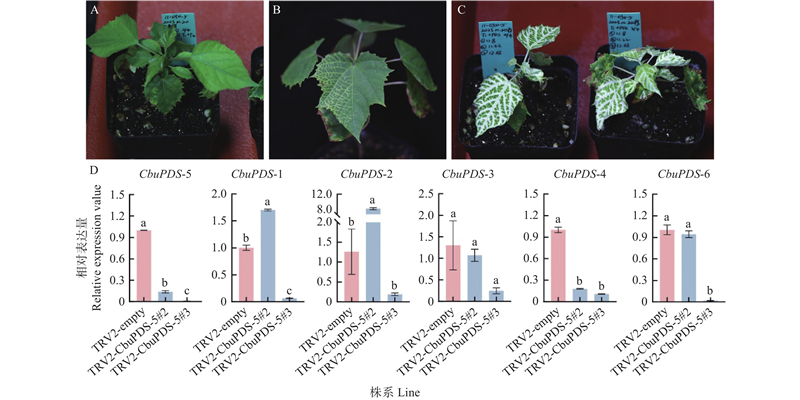
Fig.9
VIGS-mediated CbuPDS-5 silencing in Catalpa bungei A: Empty vector;B: Results of infection with only one injection;C: TRV2-CbuPDS-5#2 and TRV2-CbuPDS-5#3;D: Expression of CbuPDSs in TRV2-CbuPDS-5#2 and TRV2-CbuPDS-5#3. Different lowercase letters refer to significant difference (P<0.05)."
| 陈段芬. 2009. 中国水仙花型、花色发育基因(NTMADS1、NtMADS3、NTPDS1和NTPZDS1)的克隆与转化. 北京: 中国林业科学研究院. | |
| Chen D F. 2009. Cloning and transformation of genes related to floral shape and colour development in Narcissus tazzetta var. chinensis. Beijing: Chinese Academy of Forestry. [in Chinese] | |
|
董 静, 段秋笛, 徐艳红, 等. 长期继代培养过程中楸树愈伤组织的分化能力. 西北农业学报, 2024, 33 (2): 373- 379.
doi: 10.7606/j.issn.1004-1389.2024.02.020 |
|
|
Dong J, Duan Q D, Xu Y H, et al. Differentiation ability of Catalpa bungei callus in long term subculture. Acta Agriculturae Boreali-occidentalis Sinica, 2024, 33 (2): 373- 379.
doi: 10.7606/j.issn.1004-1389.2024.02.020 |
|
| 郝梦媛, 杭 琦, 师恭曜. VIGS基因沉默技术在作物基因功能研究中的应用与展望. 中国农业科技导报, 2022, 24 (1): 1- 13. | |
| Hao M Y, Hang Q, Shi G Y. Application and prospect of virus-induced gene silencing in crop gene function research. Journal of Agricultural Science and Technology, 2022, 24 (1): 1- 13. | |
| 何 荷, 贾瑞瑞, 付 钰, 等. 2023. 不同砧穗组合楸树嫁接苗的生理生化特性. 林业科学, 59(10): 99-112. | |
| He H, Jia R R, Fu Y, et al. 2023. Physiological and biochemical traits of grafted seedlings of Catalpa bungei with various combinations of rootstocks and scions. Scientia Silvae Sinicae, 59(10): 99-112. [in Chinese] | |
| 康向阳. 2024. 关于我国林木育种向智能分子设计育种发展的思考. 北京林业大学学报, 46(3): 1-7. | |
| Kang X Y. 2024. Thoughts on the development of forest tree breeding towards intelligent molecular design breeding in China. Journal of Beijing Forestry University, 46(3): 1−7. [in Chinese] | |
| 李芳军. 2014. 利用VIGS技术研究棉花抗逆基因功能. 北京: 中国农业大学. | |
| Li F J. 2014. Analysis function of cotton resistance genes by virus-induced gene silencing (VIGS). Beijing: China Agricultural University. [in Chinese] | |
| 王军辉. 林木新种质创制研究进展. 中国农业科技导报, 2022, 24 (12): 129- 141. | |
| Wang J H. Research progress on development of new germplasm of forest trees. Journal of Agricultural Science and Technology, 2022, 24 (12): 129- 141. | |
| 杨 莎, 张 彬, 韩 垚, 等. 矮牵牛PDS基因的克隆及其在shRNA介导的基因沉默中的应用. 园艺学报, 2017, 44 (2): 315- 322. | |
| Yang S, Zhang B, Han Y, et al. Cloning of petunia PDS gene and its application in shRNA-mediated gene silencing. Acta Horticulturae Sinica, 2017, 44 (2): 315- 322. | |
| 朱海生, 李永平, 温庆放, 等. 草莓八氢番茄红素脱氢酶基因pds的克隆及特征分析. 园艺学报, 2011, 38 (1): 55- 60. | |
| Zhu H S, Li Y P, Wen Q F, et al. Cloning and characterization of pds gene in Fragaria × ananassa. Acta Horticulturae Sinica, 2011, 38 (1): 55- 60. | |
|
Chen C J, Wu Y, Li J W, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining. Molecular Plant, 2023, 16 (11): 1733- 1742.
doi: 10.1016/j.molp.2023.09.010 |
|
|
Deng Y, Yarur-Thys A, Baulcombe D C. Virus-induced overexpression of heterologous FLOWERING LOCUS T for efficient speed breeding in tomato. Journal of Experimental Botany, 2024, 75 (1): 36- 44.
doi: 10.1093/jxb/erad369 |
|
|
Duvaud S, Gabella C, Lisacek F, et al. Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Research, 2021, 49 (w1): W216- W227.
doi: 10.1093/nar/gkab225 |
|
|
Fan D, Liu T T, Li C F, et al. Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Scientific Reports, 2015, 5, 12217.
doi: 10.1038/srep12217 |
|
|
He G R, Cao Y W, Wang J, et al. WUSCHEL-related homeobox genes cooperate with cytokinin to promote bulbil formation in Lilium lancifolium. Plant Physiology, 2022, 190 (1): 387- 402.
doi: 10.1093/plphys/kiac259 |
|
| Horton P, Park K J, Obayashi T, et al. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Research, 35(web server issue): W585–W587. | |
|
Jarugula S, Willie K, Stewart L R. Barley stripe mosaic virus (BSMV) as a virus-induced gene silencing vector in maize seedlings. Virus Genes, 2018, 54 (4): 616- 620.
doi: 10.1007/s11262-018-1569-9 |
|
| Komatsu H, Abdellatif I M Y, Yuan S Z, et al. 2020. Genome editing in PDS genes of tomatoes by non-selection method and of Nicotiana benthamiana by one single guide RNA to edit two orthologs. Plant Biotechnology (Tokyo, Japan), 37(2): 213–221. | |
| Kumagai M H, Donson J, Della-Cioppa G, et al. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proceedings of the National Academy of Sciences of the United States of America, 1995, 92 (5): 1679- 1683. | |
|
Li X R, Zuo X, Li M M, et al. Efficient CRISPR/Cas9-mediated genome editing in Rehmannia glutinosa. Plant Cell Reports, 2021, 40 (9): 1695- 1707.
doi: 10.1007/s00299-021-02723-3 |
|
|
Naing A H, Kyu S Y, Pe P P W, et al. Silencing of the phytoene desaturase (PDS) gene affects the expression of fruit-ripening genes in tomatoes. Plant Methods, 2019, 15, 110.
doi: 10.1186/s13007-019-0491-z |
|
|
Qi X Y, Mo Q P, Li J, et al. Establishment of virus-induced gene silencing (VIGS) system in Luffa acutangula using phytoene desaturase (PDS) and tendril synthesis related gene (TEN). Plant Methods, 2023, 19 (1): 94.
doi: 10.1186/s13007-023-01064-4 |
|
|
Sandmann G. Molecular evolution of carotenoid biosynthesis from bacteria to plants. Physiologia Plantarum, 2002, 116 (4): 431- 440.
doi: 10.1034/j.1399-3054.2002.1160401.x |
|
|
Senthil-Kumar M, Hema R, Anand A, et al. A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytologist, 2007, 176 (4): 782- 791.
doi: 10.1111/j.1469-8137.2007.02225.x |
|
|
Wu K X, Wu Y D, Zhang C W, et al. Simultaneous silencing of two different Arabidopsis genes with a novel virus-induced gene silencing vector. Plant Methods, 2021, 17 (1): 6.
doi: 10.1186/s13007-020-00701-6 |
|
|
Zhang M M, Liu B Y, Fei Y, et al. Genetic architecture of leaf morphology revealed by integrated trait module in Catalpa bungei. Horticulture Research, 2023, 10 (4): uhad032.
doi: 10.1093/hr/uhad032 |
| [1] | Zhao Shan, Dong Jing, Zheng Shuai, Chen Faju, Liu Wen, Liang Hongwei. Reasons for the Difficulty of Differentiation of Embryogenic Callus in Long-Term Subculture of Catalpa bungei [J]. Scientia Silvae Sinicae, 2025, 61(6): 130-138. |
| [2] | Fangyu Yin,Yamin Du,Zhu Li,Jiali Jiang. Shrinkage and Swelling Behavior of Different Types of Tissues in Catalpa bungei Wood [J]. Scientia Silvae Sinicae, 2024, 60(7): 105-116. |
| [3] | He He,Ruirui Jia,Yu Fu,Yanyan Zhu,Lianggui Wang,Xiulian Yang. Physiological and Biochemical Traits of Grafted Seedlings of Catalpa bungei with Various Combinations of Rootstocks and Scions [J]. Scientia Silvae Sinicae, 2023, 59(10): 99-112. |
| [4] | Yunxin Cen,Jia Liu,Faju Chen,Jingyuan Yang,Qiang Liu,Tao Wang,Hongwei Liang. Agrobacterium-Mediated Genetic Transformation System of Catalpa bungei [J]. Scientia Silvae Sinicae, 2021, 57(8): 195-204. |
| [5] | Bai Ouyang,Zhu Li,Jiali Jiang. Hygroscopicity and Swelling Behavior of Catalpa bungei Earlywood and Latewood [J]. Scientia Silvae Sinicae, 2021, 57(5): 176-183. |
| [6] | Xiao Yao, Yi Fei, Han Donghua, Lu Nan, Yang Guijuan, Zhao Kun, Wang Junhui, Ma Wenjun. Difference Analysis of Growth and Nitrogen Utilization and Distribution in Photosynthetic System of Catalpa bungei Intraspecific and Interspecific Hybrids [J]. Scientia Silvae Sinicae, 2019, 55(5): 55-64. |
| [7] | Zhang Enliang, Ma Lingling, Yang Rutong, Li Linfang, Wang Qing, Li Ya, Wang Peng. Transcriptome Profiling of IBA-Induced Adventitious Root Formation in Softwood Cuttings of Catalpa bungei ‘Yu-1’ [J]. Scientia Silvae Sinicae, 2018, 54(5): 48-61. |
| [8] | Mao Weibing, Chen Faju, Wang Changlan, Liang Hongwei. Transcriptome Sequencing and Analysis of Male Sterile Flower Buds in Catalpa bungei [J]. Scientia Silvae Sinicae, 2017, 53(6): 141-150. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||