

Scientia Silvae Sinicae ›› 2025, Vol. 61 ›› Issue (11): 35-44.doi: 10.11707/j.1001-7488.LYKX20240721
• Frontiers and hot topics • Previous Articles Next Articles
Hui Li,Qingnan Wang,Chenglei Zhu,Huayu Sun,Xiaolin Di,Zhimin Gao*( )
)
Received:2024-11-30
Revised:2025-04-07
Online:2025-11-25
Published:2025-12-11
Contact:
Zhimin Gao
E-mail:gaozhimin@icbr.ac.cn
CLC Number:
Hui Li,Qingnan Wang,Chenglei Zhu,Huayu Sun,Xiaolin Di,Zhimin Gao. Identification and Function of Rubisco Activase Gene Family in Three Bamboo Species[J]. Scientia Silvae Sinicae, 2025, 61(11): 35-44.
Table 2
Basic characteristics of the proteins encoded by Rcas in different bamboo species"
| 基因 Gene | 基因登录号 Accession ID | 蛋白长度 Protein length/aa | 理论等电点 Isoelectric point | 亲水性总平均值 Grand average of hydropathicity (GRAVY) | 转运肽长度 Transit peptide length/aa |
| PeRca1-β | PH02Gene18719.t1 | 427 | 6.93 | –0.376 | 48 |
| PeRca2-α | PH02Gene23790.t1 | 463 | 6.20 | –0.411 | 48 |
| PeRca3-α | PH02Gene23791.t1 | 489 | 6.15 | –0.377 | 48 |
| BamRca1-β | Bam009417.1 | 430 | 6.08 | –0.380 | 48 |
| BamRca2-α | Bam022659.1 | 465 | 5.68 | –0.439 | 47 |
| BamRca3-β | Bam031930.1 | 430 | 6.08 | –0.368 | 48 |
| OlaRca1-β | Ola008925.1 | 428 | 6.49 | –0.423 | 48 |
| OlaRca2-α | Ola008927.1 | 465 | 6.20 | –0.372 | 47 |

Fig.1
Rca gene structure analysis in different bamboo species and sequence alignment of C terminal extension in their encoding proteins A: Gene structure; B: Alignment of protein sequences (The two red boxes show the Cys residues predicted to be involved in redox regulation. Bam: Bambusa amplexicauli;Bd: Brachypodium distachyon;Hv: Hordeum vulgare;Ola: Olyra latifolia;Os: Oryza sativa;Pe: Phyllostachys edulis;Sb: Sorghum bicolor;Si: Setaria italica;Ta: Tritticum aestivum;Zm: Zea mays. α and β indicate two isoforms)."
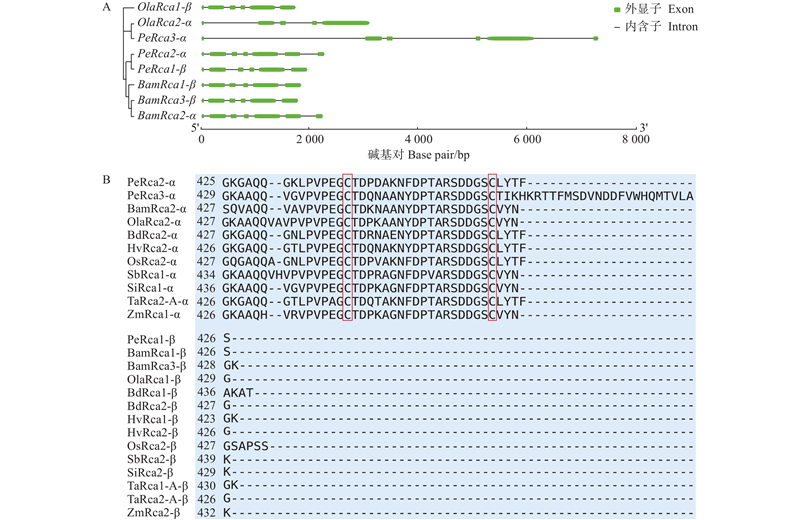
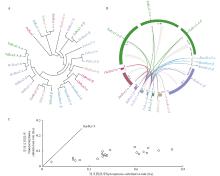
Fig.2
Evolution of Rcas in Poaceae plants A: Phylogenetic tree of Rcas in Poaceae plants;B: Collinearity analysis (Gray curves denote for all homolog pairs,and highlight curves for the homolog pairs of Rca family members between moso bamboo with other species);C: The scatter plot shows the Ka/Ks ratios of collinear pairs."
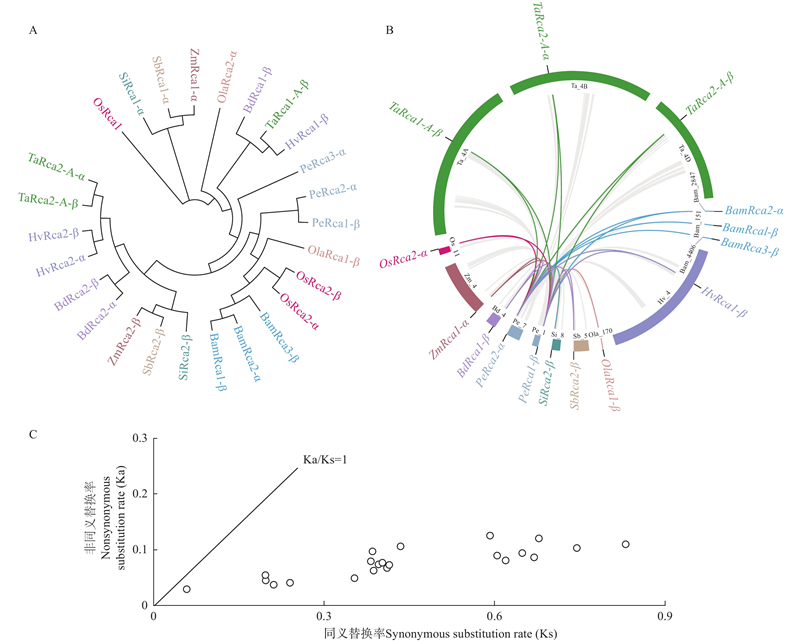
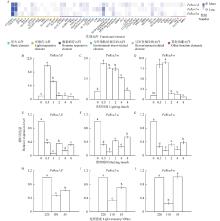
Fig.4
Cis-element classification in the promoters of PeRcas and expression levels of PeRcas under different light treatments A: Cis-element classification of PeRcas promoter;B–J: Expression levels of PeRcas (B–D: Different light times;E–F: Different dark times;H–J: Different shade treatments for 2 hours). Different lowercase letters indicate significant differences under different treatments (P<0.05)."
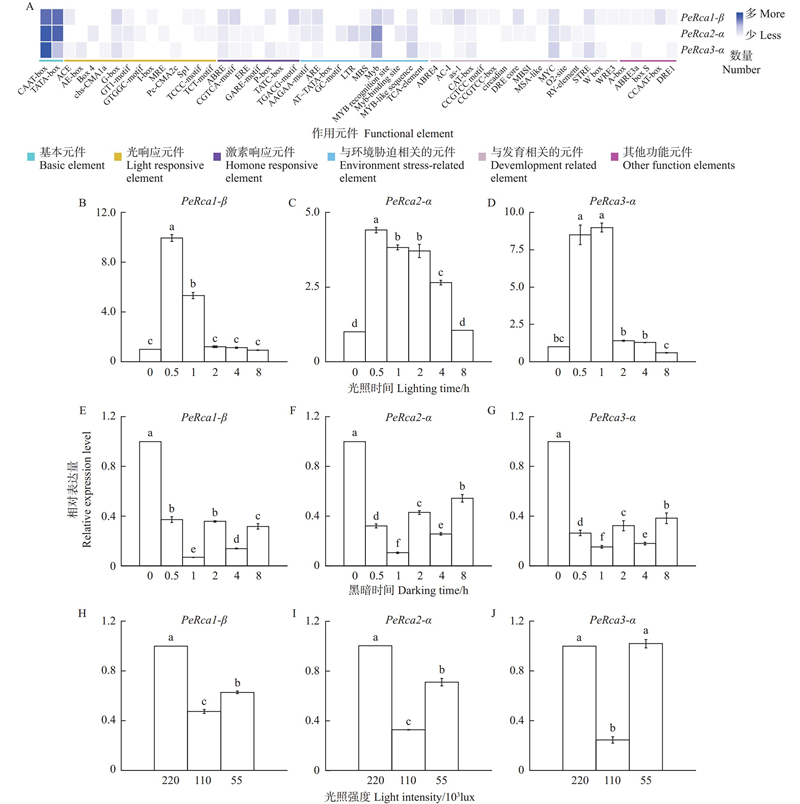
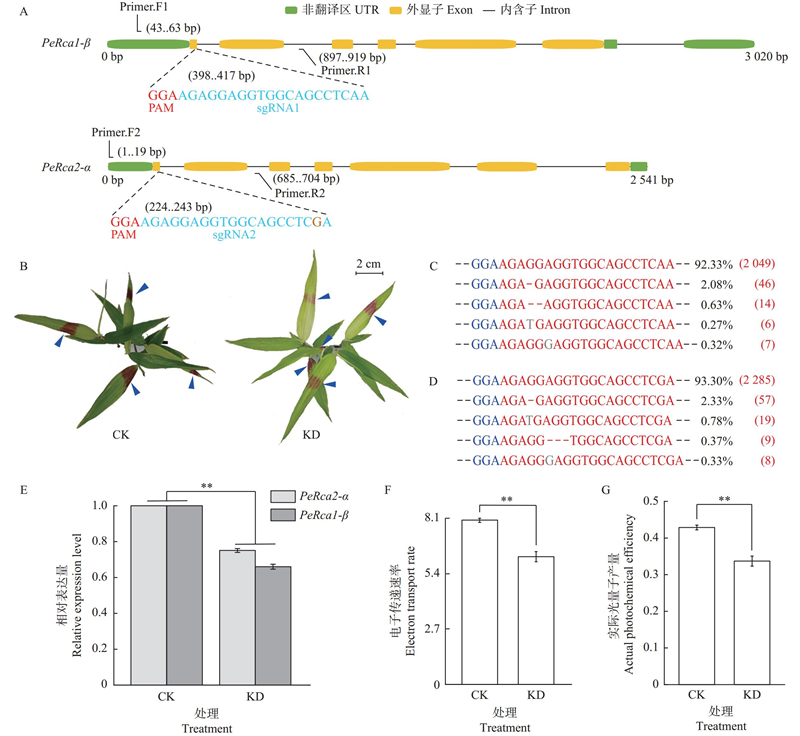
Fig.7
In-planta gene editing in moso bamboo A: Schematic diagrams of sgRNA designs of PeRca1-β and PeRca2-α (PAM: Protospacer adjacent motif;sgRNA: Small guide RNA;Primer: Primer used for amplification of target fragments);B: In-planta gene editing in moso bamboo seedlings for 7 days (The blue arrows point to the edited area, CK: control, KD: knockdown by gene editing);C, D: Deep sequencing results of the fragments generated by the edited PeRcas (Portions of sequences in red, blue, and grey indicate the target sites, PAM, and insertions, respectively. The red dashes indicate deleted nucleotides);E: The relative expression levels of PeRcas;F, G: Chlorophyll fluorescence index. Significant differences are calculated by Student’s t-tests (** P<0.01)."
| 陈丽洁, 李红艳, 黄建波, 等. 外源施硒对绿竹生长、光合作用以及硒含量的影响. 经济林研究, 2024, 42 (2): 188- 195. | |
| Chen L J, Li H Y, Huang J B, et al. Effects of exogenous selenium application on growth, photosynthesis and selenium content in Dendrocalamopsis oldhami. Non-Wood Forest Research, 2024, 42 (2): 188- 195. | |
|
黎 茵, 张以顺. 农杆菌注射渗透法转化烟草实验研究. 实验技术与管理, 2010, 27 (11): 50- 52.
doi: 10.3969/j.issn.1002-4956.2010.11.015 |
|
|
Li Y, Zhang Y S. Study on agrobacterium tumefaciens-mediated transient transformation of tobacco by infiltration. Experimental Technology and Management, 2010, 27 (11): 50- 52.
doi: 10.3969/j.issn.1002-4956.2010.11.015 |
|
|
王灵杰, 栗青丽, 高培军, 等. 毛竹茎秆快速生长期光合关键酶活性及基因表达分析. 浙江农林大学学报, 2021, 38 (1): 84- 92.
doi: 10.11833/j.issn.2095-0756.20200277 |
|
|
Wang L J, Li Q L, Gao P J, et al. Activities of key enzymes involved in photosynthesis and expression patterns of corresponding genes during rapid growth of Phyllostachys edulis. Journal of Zhejiang A&F University, 2021, 38 (1): 84- 92.
doi: 10.11833/j.issn.2095-0756.20200277 |
|
| 杨 丽, 娄永峰. 毛竹、麻竹光合途径类型分析. 南京林业大学学报(自然科学版), 2015, 39 (5): 169- 173. | |
| Yang L, Lou Y F. A preliminary study on the type of photosynthetic pathway in Phyllostachys edulis and Dendrocalamus latiflorus. Journal of Nanjing Forestry University (Natural Sciences Edition), 2015, 39 (5): 169- 173. | |
|
周国模, 姜培坤. 毛竹林的碳密度和碳贮量及其空间分布. 林业科学, 2004, 40 (6): 20- 24.
doi: 10.3321/j.issn:1001-7488.2004.06.004 |
|
|
Zhou G M, Jiang P K. Density storage and spatial distribution of carbon in Phyllostachy pubescens forest. Scientia Silvae Sinicae, 2004, 40 (6): 20- 24.
doi: 10.3321/j.issn:1001-7488.2004.06.004 |
|
|
Andersson I, Backlund A. Structure and function of rubisco. Plant Physiology and Biochemistry, 2008, 46 (3): 275- 291.
doi: 10.1016/j.plaphy.2008.01.001 |
|
|
Carmo-Silva A E, Salvucci M E. The regulatory properties of rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiology, 2013, 161 (4): 1645- 1655.
doi: 10.1104/pp.112.213348 |
|
| Chen M, Ji M L, Wen B B, et al. GOLDEN 2-LIKE transcription factors of plants. Frontiers in Plant Science, 2016, 7, 1509. | |
|
Jin S H, Wang D, Zhu F Y, et al. Up-regulation of cyclic electron flow and down-regulation of linear electron flow in antisense-rca mutant rice. Photosynthetica, 2008, 46 (4): 506- 510.
doi: 10.1007/s11099-008-0086-9 |
|
| Liu Y, Zhu C L, Lin Z M, et al. Systematic identification and validation of the reference genes from 447 transcriptome datasets of moso bamboo (Phyllostachys edulis). Horticultural Plant Journal, 2024, 11 (3): 1353- 1363. | |
|
Liu Z, Taub C C, McClung C R. Identification of an Arabidopsis thaliana ribulose-1, 5-bisphosphate carboxylase/oxygenase activase (RCA) minimal promoter regulated by light and the circadian clock. Plant Physiology, 1996, 112 (1): 43- 51.
doi: 10.1104/pp.112.1.43 |
|
|
Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods, 2001, 25 (4): 402- 408.
doi: 10.1006/meth.2001.1262 |
|
| Nagarajan R, Gill K S. Evolution of Rubisco activase gene in plants. Plant Molecular Biology, 2018, 96 (1/2): 69- 87. | |
| Pan T, Zhang F K, Sheng L, et al. Cloning and characterization of the Rubisco activase gene from Pinus massoniana. Plant Molecular Biology Reporter, 2023, 41 (1): 81- 91. | |
| Portis A R Jr, Li C S, Wang D F, et al. Regulation of rubisco activase and its interaction with rubisco. Journal of Experimental Botany, 2008, 59 (7): 1597- 1604. | |
|
Robinson S P, Portis A R. Release of the nocturnal inhibitor, caryarabinitol-1-phosphate, from ribulose bisphosphate carylase/oxygenase by rubisco activase. FEBS Letters, 1988, 233 (2): 413- 416.
doi: 10.1016/0014-5793(88)80473-3 |
|
| Salvucci M E. Potential for interactions between the carboxy- and amino-termini of Rubisco activase subunits. FEBS Letters, 2004, 560 (1/2/3): 205- 209. | |
|
Salvucci M E, Werneke J M, Ogren W L, et al. Purification and species distribution of Rubisco activase. Plant Physiology, 1987, 84 (3): 930- 936.
doi: 10.1104/pp.84.3.930 |
|
| Stotz M, Mueller-Cajar O, Ciniawsky S, et al. Structure of green-type Rubisco activase from tobacco. Nature Structural & Molecular Biology, 2011, 18 (12): 1366- 1370. | |
|
Sun H Y, Lou Y F, Li H, et al. Unveiling the intrinsic mechanism of photoprotection in bamboo under high light. Industrial Crops and Products, 2024, 209, 118049.
doi: 10.1016/j.indcrop.2024.118049 |
|
|
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 2021, 38 (7): 3022- 3027.
doi: 10.1093/molbev/msab120 |
|
|
Wu H R, Li L B, Jing Y X, et al. Over-and anti-sense expressions of the large isoform of ribulose-1, 5-bisphosphate carboxylase/oxygenase activase gene in Oryza sativa affect the photosynthetic capacity. Photosynthetica, 2007, 45 (2): 194- 201.
doi: 10.1007/s11099-007-0032-2 |
|
|
Yang L, Lou Y F, Peng Z H, et al. Molecular characterization and primary functional analysis of PeMPEC, a magnesium-protoporphyrin IX monomethyl ester cyclase gene of bamboo (Phyllostachys edulis). Plant Cell Reports, 2015, 34 (11): 2001- 2011.
doi: 10.1007/s00299-015-1846-1 |
| [1] | Rui Gu,Shaohui Fan,Songpo Wei,Guanglu Liu. Construction of a Core Germplasm of Moso Bamboo Based on Phenotypic Traits [J]. Scientia Silvae Sinicae, 2025, 61(9): 101-112. |
| [2] | Yanting Chang,Tao Hu,Xue Zhang,Zehui Jiang,Yanjun Ma,Yayun Deng,Wenbo Zhang. Cloning and Function of PoLEC1 Gene in Paeonia ostii [J]. Scientia Silvae Sinicae, 2025, 61(9): 138-145. |
| [3] | Jiangfei Wang,Hui Li,Chenglei Zhu,Xiaolin Di,Ying Li,Qingnan Wang,Huiru Wan,Huayu Sun,Zhimin Gao. Functions of PeBAM3 of Moso Bamboo Involved in Leaf Starch Degradation [J]. Scientia Silvae Sinicae, 2025, 61(8): 231-240. |
| [4] | Xiaoling Yan,Qin Hao,Zi Shen,Yujia Zhang,Xiaoqin Guo. Expression, Protein Interaction and Biological Function Analysis of PheFT1 Gene in Moso Bamboo [J]. Scientia Silvae Sinicae, 2025, 61(4): 140-152. |
| [5] | Xinxin Ma,You Wang,Jiajun Wang,Long Feng,Jianfeng Ma. Changes in Ash Composition of Bamboo during Pyrolysis and the Distribution Pattern of Silicon Transformation [J]. Scientia Silvae Sinicae, 2025, 61(2): 172-179. |
| [6] | Ao Zhang,Wenting Li,Tianxiang Wang,Yaoxing Wu,Gang Lei,Lianghua Qi. Regional Differentiation and It’s Influencing Factors of Soil Easily-oxidized Organic Carbon in Subtropical Phyllostachys edulis Forests [J]. Scientia Silvae Sinicae, 2024, 60(6): 1-12. |
| [7] | Yi Wang,Junwei Luan,Chen Chen,Shirong Liu. Asymmetric Response of Soil Respiration and Its Components to Nitrogen and Phosphorus Addition in Phyllostachys edulis Forest [J]. Scientia Silvae Sinicae, 2023, 59(7): 54-64. |
| [8] | Jinling Yuan,Jinjun Yue,Jingxia Ma,Lei Yu,Lei Liu. Culm Form Characteristics of Phyllostachys edulis ‘Yuanbao’ [J]. Scientia Silvae Sinicae, 2023, 59(5): 71-80. |
| [9] | Wei Zhang,Yuyou He,Ziwu Guo,Sheping Wang,Shuanglin Chen. Characteristics of Arbor Species Community Structure and Diversity in the Succession of Out-of-Management Phyllostachys edulis Forest [J]. Scientia Silvae Sinicae, 2022, 58(12): 12-20. |
| [10] | Jiamin Xie,Mingbing Zhou. Identification and Bioinformatics Analysis of Mariner-Like Element Autonomous Transposons in Phyllostachys edulis [J]. Scientia Silvae Sinicae, 2022, 58(1): 175-184. |
| [11] | Minhao Liu,Long Li,Jing Ye,Xuanyuan Zhou,Zhouqi Li,Ruishen Fan,Junlei Xu. Genome-Wide Identification and Expression Analysis of the ARF Gene Family in Eucommia ulmoides [J]. Scientia Silvae Sinicae, 2021, 57(3): 170-180. |
| [12] | Wenjie Hu,Hongdong Pang,Xingyi Hu,Faxin Huang,Jiawei Yang,Lijun Xu,Miao Gong. Effects of Bamboo Forest Density and Fertilizer Types on the Yield and Quality of Phyllostachys edulis Bamboo Shoots and Soil Physicochemical Properties in Mufu Mountain Area [J]. Scientia Silvae Sinicae, 2021, 57(12): 32-42. |
| [13] | Zhaoli Chen,Niu Yu,Rongsheng Li,Wentao Zou,Mingliang Dong,Maocheng Zhu,Jinchang Yang. Cloning and Expression Analysis of Sesquiterpene Synthase Gene SgSTPS3 in Sindora glabra [J]. Scientia Silvae Sinicae, 2021, 57(11): 68-78. |
| [14] | Chenglei Zhu,Kebin Yang,Xiurong Xu,Shuang Ma,Xiaopei Li,Zhimin Gao. Molecular Characteristics of NIP Genes in Phyllostachys edulis and Their Expression Patterns in Response to Stresses [J]. Scientia Silvae Sinicae, 2021, 57(1): 64-76. |
| [15] | Linxin Fang,Shouke Zhang,Kefeng Jia,Bihuan Ye,Wei Zhang,Jinping Shu,Haojie Wang,Tiansen Xu. Oviposition Preference of Eutomostethus deqingensis (Hymenoptera: Tenthredinidae) on Phyllostachys edulis [J]. Scientia Silvae Sinicae, 2021, 57(1): 131-139. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||