

Scientia Silvae Sinicae ›› 2020, Vol. 56 ›› Issue (4): 74-81.doi: 10.11707/j.1001-7488.20200408
Special Issue: 林木育种
• Articles • Previous Articles Next Articles
Peihuang Zhu,Yu Chen,Lingzhi Zhu,Rong Li,Kongshu Ji
Received:2019-03-18
Online:2020-04-25
Published:2020-05-29
CLC Number:
Peihuang Zhu,Yu Chen,Lingzhi Zhu,Rong Li,Kongshu Ji. Codon Usage Bias and Its Influencing Factors in Pinus massoniana Transcriptome[J]. Scientia Silvae Sinicae, 2020, 56(4): 74-81.
Table 1
Putative optimal codons in the transcriptome of P. massoniana"
| 氨基酸 Amino acid | 密码子 Codon | 低表达基因 Low expressed genes | 高表达基因 High expressed genes | ΔRSCU | |||
| RSCU | 出现次数 Number | RSCU | 出现次数 Number | ||||
| Phe | TTT* | 1.09 | 1 452 | 1.32 | 1 381 | 0.23 | |
| TTC | 0.91 | 1 180 | 0.68 | 830 | -0.23 | ||
| Leu | TTA* | 0.77 | 711 | 1.05 | 917 | 0.28 | |
| TTG* | 1.38 | 1 245 | 1.67 | 1 363 | 0.29 | ||
| CTT* | 1.21 | 1 120 | 1.42 | 1 136 | 0.21 | ||
| CTC | 0.92 | 890 | 0.51 | 440 | -0.41 | ||
| CTA | 0.56 | 544 | 0.56 | 358 | 0 | ||
| CTG | 1.17 | 1 084 | 0.79 | 614 | -0.38 | ||
| Ile | ATT* | 1.35 | 1 410 | 1.60 | 1 704 | 0.25 | |
| ATC | 0.82 | 878 | 0.59 | 703 | -0.23 | ||
| ATA | 0.83 | 872 | 0.80 | 700 | -0.03 | ||
| Met | ATG | 1.00 | 1 737 | 1.00 | 1 569 | 0 | |
| Val | GTT* | 1.36 | 1 369 | 1.69 | 1 615 | 0.33 | |
| GTC | 0.74 | 819 | 0.53 | 467 | -0.21 | ||
| GTA* | 0.72 | 730 | 0.82 | 716 | 0.10 | ||
| GTG | 1.18 | 1 220 | 0.96 | 895 | -0.22 | ||
| Tyr | TAT* | 1.16 | 950 | 1.43 | 1 165 | 0.27 | |
| TAC | 0.84 | 740 | 0.57 | 529 | -0.27 | ||
| TER | TAA | 1.04 | 105 | 1.20 | 133 | 0.16 | |
| TAG | 0.66 | 58 | 0.59 | 53 | -0.07 | ||
| His | CAT* | 1.18 | 804 | 1.49 | 1 000 | 0.31 | |
| CAC | 0.82 | 600 | 0.51 | 361 | -0.31 | ||
| Gln | CAA* | 0.91 | 1 129 | 1.14 | 1 446 | 0.23 | |
| CAG | 1.09 | 1 370 | 0.86 | 880 | -0.23 | ||
| Asn | AAT* | 1.22 | 1 616 | 1.43 | 1 751 | 0.21 | |
| AAC | 0.78 | 1 061 | 0.57 | 789 | -0.21 | ||
| Lys | AAA* | 0.94 | 1 887 | 1.07 | 2 250 | 0.13 | |
| AAG | 1.06 | 2 155 | 0.93 | 1 755 | -0.13 | ||
| Asp | GAT* | 1.27 | 2 031 | 1.49 | 2 041 | 0.22 | |
| GAC | 0.73 | 1 131 | 0.51 | 739 | -0.22 | ||
| Glu | GAA* | 1.06 | 2 196 | 1.22 | 2 264 | 0.16 | |
| GAG | 0.94 | 1 943 | 0.78 | 1 346 | -0.16 | ||
| Ser | TCT* | 1.33 | 1 185 | 1.78 | 1 300 | 0.45 | |
| TCC | 0.96 | 909 | 0.59 | 478 | -0.37 | ||
| TCA* | 1.11 | 973 | 1.67 | 1 217 | 0.56 | ||
| TCG | 0.73 | 683 | 0.25 | 211 | -0.48 | ||
| Pro | CCT* | 1.35 | 1 013 | 1.65 | 1 148 | 0.30 | |
| CCC | 0.88 | 669 | 0.47 | 372 | -0.41 | ||
| CCA* | 1.14 | 866 | 1.65 | 1 198 | 0.51 | ||
| CCG | 0.62 | 508 | 0.23 | 172 | -0.39 | ||
| Thr | ACT* | 1.20 | 878 | 1.53 | 1 088 | 0.33 | |
| ACC | 0.87 | 698 | 0.57 | 467 | -0.30 | ||
| ACA* | 1.30 | 911 | 1.65 | 1 163 | 0.35 | ||
| ACG | 0.63 | 474 | 0.25 | 175 | -0.38 | ||
| Ala | GCT* | 1.24 | 1 420 | 1.64 | 1 893 | 0.40 | |
| GCC | 0.92 | 1 123 | 0.53 | 670 | -0.39 | ||
| GCA* | 1.27 | 1 450 | 1.62 | 1 489 | 0.35 | ||
| GCG | 0.57 | 692 | 0.21 | 260 | -0.36 | ||
| Cys | TGT* | 0.97 | 605 | 1.27 | 621 | 0.30 | |
| TGC | 1.03 | 665 | 0.73 | 310 | -0.30 | ||
| TER | TGA | 1.30 | 117 | 1.21 | 94 | -0.09 | |
| Trp | TGG | 1.00 | 781 | 1.00 | 680 | 0 | |
| Arg | CGT* | 0.76 | 466 | 0.86 | 566 | 0.10 | |
| CGC | 0.76 | 497 | 0.33 | 154 | -0.43 | ||
| CGA | 0.80 | 462 | 0.67 | 406 | -0.13 | ||
| CGG | 0.73 | 466 | 0.41 | 160 | -0.32 | ||
| Ser | AGT* | 0.90 | 821 | 1.08 | 804 | 0.18 | |
| AGC | 0.98 | 875 | 0.62 | 448 | -0.36 | ||
| Arg | AGA* | 1.53 | 914 | 2.15 | 1 157 | 0.62 | |
| AGG* | 1.41 | 866 | 1.58 | 668 | 0.17 | ||
| Gly | GGT* | 0.99 | 1 043 | 1.32 | 1 606 | 0.33 | |
| GGC | 0.89 | 976 | 0.61 | 615 | -0.28 | ||
| GGA* | 1.23 | 1 262 | 1.38 | 1 451 | 0.15 | ||
| GGG | 0.89 | 939 | 0.68 | 577 | -0.21 | ||
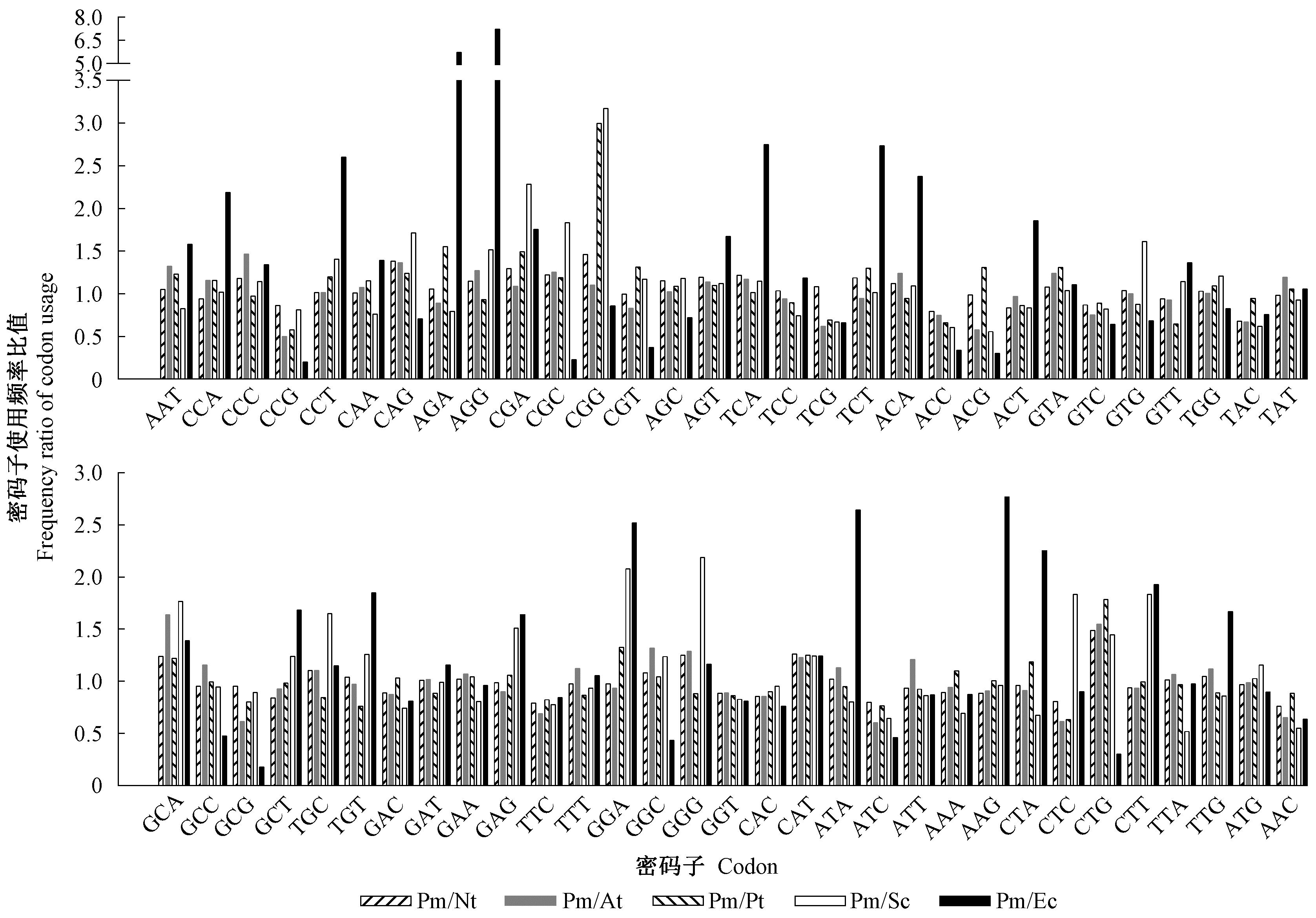
Fig.4
Comparison of frequency ratio of codon usage between P. massoniana transcriptome and representative organism genomes Pm/Nt, Pm/At, Pm/Pt, Pm/Sc and Pm/Ec indicate frequency ratio of codon usage between Pinus massoniana transcriptome and genomes of Nicotiana tabacum, Arabidopsis thaliana, Populus tremula, Saccharomyces cerevisiae and Escherichia coli."
| 蔡海莺, 李杨, 张辉, 等. 基因设计对重组蛋白表达的影响研究进展. 生物工程学报, 2013. 29 (9): 1201- 1213. | |
| Cai H Y , Li Y , Zhang H , et al. Effects of gene design on recombinant protein expression. Chinese Journal of Biotechnology, 2013. 29 (9): 1201- 1213. | |
| 陈天华, 张若思, 姜国珍, 等. 产蒎烯人工酵母细胞的构建. 化工学报, 2019. 70 (1): 179- 188. | |
| Chen T H , Zhang R S , Jiang G Z , et al. Metabolic engineering of Saccharomyces cerevisiae for pinene production. CIESC Journal, 2019. 70 (1): 179- 188. | |
| 陈晓明, 陈博雯, 李魁鹏, 等. 马尾松GGPPS基因与产脂力相关性分析. 分子植物育种, 2018. 16 (16): 5247- 5254. | |
| Chen X M , Chen B W , Li K P , et al. Correlation analysis of GGPPS gene and resin producing capacity in Pinus massoniana. Molecular Plant Breeding, 2018. 16 (16): 5247- 5254. | |
| 陈哲, 胡福初, 王祥和, 等. 菠萝密码子使用偏好性分析. 果树学报, 2017. 34 (8): 946- 955. | |
| Chen Z , Hu F C , Wang X H , et al. Analysis of codon usage bias of Ananas comosus with genome sequencing data. Journal of Fruit Science, 2017. 34 (8): 946- 955. | |
| 范三红, 郭蔼光, 单丽伟, 等. 拟南芥基因密码子偏爱性分析. 生物化学与生物物理进展, 2003. 30 (2): 221- 225. | |
| Fan S H , Guo A G , Shan L W , et al. Analysis of genetic code preference in Arabidopsis thaliana. Progress in Biochemistry and Biophysics, 2003. 30 (2): 221- 225. | |
| 冯红茹, 杨建明, 秦利, 等. β-蒎烯合成酶(QH6)在大肠杆菌中的表达及其产β-蒎烯的研究. 生物加工过程, 2015. 13 (1): 28- 34, 46. | |
| Feng H R , Yang J M , Qin L , et al. Expression of β-pinene synthase(QH6) in Escherichia coli for the biosynthesis of β-pinene. Chinese Journal of Bioprocess Engineering, 2015. 13 (1): 28- 34, 46. | |
| 郭天玮, 苏江, 阮维程, 等. 马尾松FLO/LFY基因的克隆及其功能分析. 分子植物育种, 2015. 13 (10): 2320- 2332. | |
| Guo T W , Su J , Ruan W C , et al. The clone and function analysis of the FLO/LFY genes in Pinus massoniana. Molecular Plant Breeding, 2015. 13 (10): 2320- 2332. | |
| 胡福初, 陈哲, 郭利军, 等. 基于RNA-Seq的莲雾密码子偏好性分析. 分子植物育种, 2017. 15 (12): 4947- 4954. | |
| Hu F C , Chen Z , Guo L J , et al. Analysis of Syzygium samarangense Merr.et Perry codon preference base on RNA-Seq. Molecular Plant Breeding, 2017. 15 (12): 4947- 4954. | |
| 寇莹莹, 宋英今, 杨少辉, 等. 植酸酶phyA基因的密码子优化及其在大豆中的表达. 作物学报, 2016. 42 (12): 1798- 1804. | |
| Kou Y Y , Song Y J , Yang S H , et al. Codon optimization and expression of phyA gene in soybean (Glycine max Merr.). Acta Agronomica Sinica, 2016. 42 (12): 1798- 1804. | |
| 赖瑞联, 冯新, 陈瑾, 等. 橄榄转录组密码子使用偏好性及其影响因素. 核农学报, 2019. 33 (1): 31- 38. | |
| Lai R L , Feng X , Chen J , et al. Codon usage bias of Canarium album (Lour.) R. transcriptome and its influence factors. Journal of Nuclear Agricultural Sciences, 2019. 33 (1): 31- 38. | |
| 李慧娟, 潘思皓, 杜函圳, 等. 川芎转录组密码子使用偏好性分析. 湖北农业科学, 2017. 56 (18): 3549- 3553. | |
| Li H J , Pan S H , Du H Z , et al. Preference analysis of codon usage of Ligusticum chuanxiong transcriptomes. Hubei Agricultural Sciences, 2017. 56 (18): 3549- 3553. | |
| 李慧敏, 谢婉凤, 冯丽贞, 等. PmACRE基因的克隆及遗传转化拟南芥. 森林与环境学报, 2018. 38 (1): 13- 19. | |
| Li H M , Xie W F , Feng L Z , et al. Isolation of PmACRE gene and its transformation in Arabidopsis thaliana. Journal of Forest and Environment, 2018. 38 (1): 13- 19. | |
| 刘慧, 王梦醒, 岳文杰, 等. 糜子叶绿体基因组密码子使用偏性的分析. 植物科学学报, 2017. 35 (3): 362- 371. | |
| Liu H , Wang M X , Yue W J , et al. Analysis of codon usage in the chloroplast genome of Broomcorn millet (Panicum miliaceum L.). Plant Science Journal, 2017. 35 (3): 362- 371. | |
| 刘慧敏, 乌云塔娜, 杜红岩. 杜仲转录组基因密码子使用偏好性分析. 北方园艺, 2016. (13): 85- 89. | |
| Liu H M , WuYun T N , Du H Y . Analysis of characteristic of codon usage of Eucommia ulmoides transcriptome. Northern Horticulture, 2016. (13): 85- 89. | |
| 刘宁, 姚元锋, 郭婧, 等. 红豆杉属基因的密码子偏性分析. 湖北农业科学, 2013. 52 (10): 2427- 2430, 2435. | |
| Liu N , Yao Y F , Guo J , et al. Analysis on codon usage bias of Taxus genes. Hubei Agricultural Sciences, 2013. 52 (10): 2427- 2430, 2435. | |
| 陆育生, 彭程, 陈喆, 等. 黄皮转录组密码子使用偏好性分析. 分子植物育种, 2018. 16 (18): 5904- 5913. | |
| Lu Y S , Peng C , Chen Z , et al. Analysis of codon usage bias in Clausena lansium transcriptome. Molecular Plant Breeding, 2018. 16 (18): 5904- 5913. | |
| 罗群凤, 冯源恒, 贾婕, 等. 马尾松叶绿体基因组测序及特征分析. 广西林业科学, 2018. 47 (4): 396- 402. | |
| Luo Q F , Feng Y H , Jia J , et al. Sequencing and characteristics analysis of chloroplast genome of Pinus massoniana. Guangxi Forestry Science, 2018. 47 (4): 396- 402. | |
| 潘婷, 张逢凯, 王晓锋, 等. 马尾松RCA基因的克隆和序列分析. 分子植物育种, 2015. 13 (2): 391- 400. | |
| Pan T , Zhang F K , Wang X F , et al. The cloning and sequence analysis of RCA genes from Pinus massoniana. Molecular Plant Breeding, 2015. 13 (2): 391- 400. | |
| 曲俊杰, 黄羽, 卢江, 等. 香蕉基因组密码子使用偏好性分析. 南方农业学报, 2017. 48 (1): 14- 19. | |
| Qu J J , Huang Y , Lu J , et al. Codon usage bias of banana genome. Journal of Southern Agriculture, 2017. 48 (1): 14- 19. | |
| 王晓锋, 何卫龙, 蔡卫佳, 等. 马尾松转录组测序和分析. 分子植物育种, 2013. 11 (3): 385- 392. | |
| Wang X F , He W L , Cai W J , et al. Analysis on transcriptome sequenced for Pinus massoniana. Molecular Plant Breeding, 2013. 11 (3): 385- 392. | |
| 王颖, 刘振宇, 吕全, 等. 马尾松α-蒎烯合成酶基因cDNA全长克隆及序列分析. 安徽农业科学, 2014. 42 (13): 3808- 3811. | |
| Wang Y , Liu Z Y , Lü Q , et al. Cloning and sequence analysis of α-pinene synthase gene from Pinus massoniana. Journal of Anhui Agriculture Sciences, 2014. 42 (13): 3808- 3811. | |
| 吴东山, 杨章旗, 黄永利. 马尾松不同半同胞家系产脂力、松脂组分分析与评价. 北京林业大学学报, 2019. 41 (2): 53- 61. | |
| Wu D S , Yang Z Q , Huang Y L . Analysis and evaluation of resin productivity and resin component among different half sibling families of Pinus massoniana. Jonrnal of Beijing Forestry University, 2019. 41 (2): 53- 61. | |
| 吴晓刚, 陈佩珍, 韦蔷, 等. 马尾松PmCOMT基因的克隆及实时定量表达分析. 分子植物育种, 2019. 17 (3): 789- 795. | |
| Wu X G , Chen P Z , Wei Q , et al. Cloning and real-time quantitative expression analysis on PmCOMT gene from Pinus massoniana. Molecular Plant Breeding, 2019. 17 (3): 789- 795. | |
| 杨章旗, 舒文波. 马尾松纸浆材育种研究进展及策略. 广西林业科学, 2012. 41 (1): 27- 32. | |
| Yang Z Q , Shu W B . Breeding progress and strategies for pulpwood of Pinus massoniana. Guangxi Forestry Science, 2012. 41 (1): 27- 32. | |
| 杨章旗. 马尾松高产脂遗传改良研究进展及育种策略. 广西林业科学, 2015. 44 (4): 317- 324. | |
| Yang Z Q . Research progress of high rosin genetic improvement and breeding strategy of Pinus massoniana. Guangxi Forestry Science, 2015. 44 (4): 317- 324. | |
| 叶友菊, 倪州献, 白天道, 等. 马尾松叶绿体基因组密码子偏好性分析. 基因组学与应用生物学, 2018. 37 (10): 4464- 4471. | |
| Ye Y J , Ni Z X , Bai T D , et al. The analysis of chloroplast genome codon usage bais in Pinus massoniana. Genomics and Applied Biology, 2018. 37 (10): 4464- 4471. | |
| 张逢凯, 潘婷, 盛璐, 等. 马尾松CAD基因的克隆及其编码蛋白质的结构预测. 分子植物育种, 2014. 12 (4): 638- 645. | |
| Zhang F K , Pan T , Sheng L , et al. Cloning and coding protein structure prediction on cDNA encoding cinnamyl alcohol dehydrogenase from Pinus massoniana. Molecular Plant Breeding, 2014. 12 (4): 638- 645. | |
| 张太奎, 起国海, 叶红莲, 等. 石榴转录组密码子使用偏向性. 园艺学报, 2017. 44 (4): 675- 690. | |
| Zhang T K , Qi G H , Ye H L , et al. Codon usage bias in pomegranate transcriptome. Acta Horticulturae Sinica, 2017. 44 (4): 675- 690. | |
| Jabeen R , Khan M S , Zafar Y , et al. Codon optimization of cry1Ab gene for hyper expression in plant organelles. Molecular Biology Reports, 2010. 37 (2): 1011- 1017. | |
| Jiang Y , Deng F , Wang H , et al. An extensive analysis on the global codon usage pattern of baculoviruses. Archives of Virology, 2008. 153 (12): 2273- 2282. | |
| Murray E E , Lotzer J , Eberle M . Codon usage in plant genes. Nucleic Acids Research, 1989. 17 (2): 477- 498. | |
| Qin Z , Cai Z , Xia G , et al. Synonymous codon usage bias is correlative to intron number and shows disequilibrium among exons in plants. BMC Genomics, 2013. 14 (1): 56. | |
| Sharp P M , Matassi G . Codon usage and genome evolution. Current Opinion in Genetics & Development, 1994. 4 (6): 851- 860. | |
| Wright F . The "effective number of codons" used in a gene. Gene, 1990. 87 (1): 23- 29. |
| [1] | Xiaorong Wang,Lei Lei,Tian Fu,Lei Pan,Lixiong Zeng,Wenfa Xiao. Short-Term Effects of Selective Cutting for Tending on Leaf Litter Decomposition Rate and Nutrient Release in Pinus massoniana Forests [J]. Scientia Silvae Sinicae, 2020, 56(4): 12-21. |
| [2] | Xing Wu,Xingfeng Hu,Peizhen Chen,Xiaobo Sun,Fan Wu,Kongshu Ji. Cloning and Functional Analysis of PmPIN1 Gene from Pinus massoniana [J]. Scientia Silvae Sinicae, 2020, 56(3): 184-192. |
| [3] | Xiaoli Yan,Wenjia Hu,Yuanfan Ma,yufan Huo,Tuo Wang,Xiangqing Ma. Nitrogen Uptake Preference of Cunninghamia lanceolata, Pinus massoniana, and Schima superba under Heterogeneous Nitrogen Supply Environment and their Root Foraging Strategies [J]. Scientia Silvae Sinicae, 2020, 56(2): 1-11. |
| [4] | Xiaoyu Lu, Zhu Chen, Fei Tang, Songling Fu, Jie Ren. Combined Transcriptomic and Metabolomic Analysis Reveals Mechanism of Anthocyanin Changes in Red Maple(Acer rubrum) Leaves [J]. Scientia Silvae Sinicae, 2020, 56(1): 38-54. |
| [5] | Zhao Qingquan, Chi Yujie, Zhang Jian, Feng Lianrong. Transcriptome Construction and Related Gene Expression Analysis of Lenzites gibbosa in Woody Environment [J]. Scientia Silvae Sinicae, 2019, 55(8): 95-105. |
| [6] | Deng Xiuxiu, Xiao Wenfa, Zeng Lixiong, Lei Lei, Shi Zheng. Transport and Distribution Characteristics of Photosynthates of Pinus massoniana Seedlings [J]. Scientia Silvae Sinicae, 2019, 55(7): 27-34. |
| [7] | Jin Guoqing, Zhang Zhen, Yu Qixin, Feng Suiqi, Feng Zhongping, Zhao Shirong, Zhou Zhichun. Comparisons of Genetic Variation and Gains of 6-year-old Families from First-and Second-Generation Seed Orchards of Pinus massoniana [J]. Scientia Silvae Sinicae, 2019, 55(7): 57-67. |
| [8] | Han Xiaohong, Lu Ciding, Hua Yin, Lin Haoyu, Shi Yufei, Wu Songqing, Zhang Feiping, Liang Guanghong. Phylogenetic Analysis of Transcriptome and Three Detoxification Enzyme Families Related Genes in Anoplophora chinensis (Coleoptera: Cerambycidae) [J]. Scientia Silvae Sinicae, 2019, 55(5): 104-113. |
| [9] | Jinchi Wang,Qinglin Huang,Zhibo Ma,Ruchu Huang,Qunrui Zheng. Species Composition and Diversity of Semi-Natural Mixed Forest of Pinus massoniana and Broad-Leaved Trees in Yong'an [J]. Scientia Silvae Sinicae, 2019, 55(11): 19-26. |
| [10] | Luo Xiaoman, Ding Guijie, Wang Yi. Active Constituents of Soil Extracts from Mycorrhizal Seedling Rhizosphere of Pinus massoniana and Their Effects on Seed Germination [J]. Scientia Silvae Sinicae, 2018, 54(8): 32-38. |
| [11] | Yao Hongyu, Liu Yamin, Zhang Shengnan, Liu Yumin, Zhou Wenying, Wang Zhenzhen. Effects of Exogenous Citric Acid on Physiological Characteristics of Pinus massoniana under Aluminum Stress [J]. Scientia Silvae Sinicae, 2018, 54(7): 155-164. |
| [12] | Zhang Enliang, Ma Lingling, Yang Rutong, Li Linfang, Wang Qing, Li Ya, Wang Peng. Transcriptome Profiling of IBA-Induced Adventitious Root Formation in Softwood Cuttings of Catalpa bungei ‘Yu-1’ [J]. Scientia Silvae Sinicae, 2018, 54(5): 48-61. |
| [13] | Wei Yongcheng, Liu Qinghua, Zhou Zhichun, Xu Liuyi, Chen Xuelian, Hao Yanping. Changes of Chemical Signals in Needles of Pinus massoniana with Different Resistance after Inoculation of Pine Wood Nematode [J]. Scientia Silvae Sinicae, 2018, 54(2): 119-125. |
| [14] | Gu Xirong, Ni Yalan, Jiang Yanan, Jia Hao. Effects of Laccaria bicolor on Growth, Uptake and Distribution of Nutrients and Aluminum of Pinus massoniana Seedlings under Acid Aluminum Exposure [J]. Scientia Silvae Sinicae, 2018, 54(2): 170-178. |
| [15] | Yin Huanhuan, Liu Qinghua, Zhou Zhichun, Yu Qixin, Feng Zhongping. Genetic Variation among Clones of Masson Pine (Pinus massoniana) for Growth, Oleoresin traits and Their Correlations [J]. Scientia Silvae Sinicae, 2018, 54(12): 82-91. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||