

林业科学 ›› 2025, Vol. 61 ›› Issue (6): 109-119.doi: 10.11707/j.1001-7488.LYKX20240401
收稿日期:2024-06-28
出版日期:2025-06-10
发布日期:2025-06-26
通讯作者:
栾启福
E-mail:qifu.luan@caf.ac.cn
基金资助:
Yadi Wu1,2,Shu Diao1,Xianyin Ding1,Qinyun Huang1,Qifu Luan1,*( )
)
Received:2024-06-28
Online:2025-06-10
Published:2025-06-26
Contact:
Qifu Luan
E-mail:qifu.luan@caf.ac.cn
摘要:
目的: 由于国外松在苗期的形态特征十分相似,区分这些良种具有一定的挑战性。本研究旨在构建一套针对国外松良种的DNA指纹图谱,为国外松种质资源管理、良种知识产权保护等提供技术支持。方法: 本研究以38份国外松本种及其杂种松良种为材料,利用湿地松和火炬松的51K液相探针芯片进行SNPs捕获,获得38个国外松改良品种的基因型数据。通过次要等位基因频率、缺失率、杂合率和多态性信息含量等遗传参数筛选能够高效识别国外松种质资源的核心SNPs;根据PIC值进一步简化SNP标记的数量,达到较少数量的SNPs能高效识样本的目的。结果: 通过SNP位点捕获和基因分型,共得到560,567个SNPs位点,筛选得到344个核心SNPs,并利用PIC值进一步精简为20个SNPs,能够高效区分湿地松、火炬松、加勒比松及其杂交松,成功建立了38个松树良种的DNA指纹图谱。结论: 开发了一套针对国外松良种的DNA指纹图谱,建立了指纹图谱鉴定国外松及其良种的方法,可为良种知识产权保护、种质管理和系谱分析提供技术支持。
中图分类号:
吴亚荻,刁姝,丁显印,黄琴韵,栾启福. 基于51K液相探针构建国外松良种的DNA指纹图谱[J]. 林业科学, 2025, 61(6): 109-119.
Yadi Wu,Shu Diao,Xianyin Ding,Qinyun Huang,Qifu Luan. Construction of a DNA Fingerprinting of Elite Varieties of Introduced Exotic Pines in China Based on 51K Liquid-Phased Probes[J]. Scientia Silvae Sinicae, 2025, 61(6): 109-119.
表2
核心SNPs的筛选步骤、条件及筛选后剩余的SNPs数量"
| 筛选步骤 Screening steps | 筛选条件 Screening criteria | 剩余SNP的数量 Number of remaining SNPs |
| 原始数据 Raw data | — | 183 849 |
| 第1步 Step 1 | 次要等位基因频率大于0.3,缺失率为0 MAF >0.3,miss rate=0 | 22 593 |
| 第2步 Step 2 | 符合哈迪-温伯格平衡 HWE <1E-4 | 13 931 |
| 第3步 Step 3 | 连锁不平衡大于0.2 LD >0.2 | 8 502 |
| 第4步 Step 4 | 多态信息含量大于0.35 PIC >0.35 | 8 502 |
| 第5步 Step 5 | 杂合度小于0.25 He <0.25 | 344 |
表4
经过数量精简后的20个SNP及其遗传多样性指数"
| 序号 No. | SNP名称 SNP ID | 染色体 Chromosome | 等位基因A Alley gene A | 等位基因B Alley gene B | 次要等位基 因频率 Minor allele frequency (MAF) | 多态信息含量 Polymorphic information content(PIC) |
| 1 | super2776.88036 | 10 | G | A | 0.500 0 | 0.500 0 |
| 2 | scaffold121890.3772 | 6 | T | C | 0.500 0 | 0.500 0 |
| 3 | scaffold124124.14341 | 11 | C | T | 0.500 0 | 0.500 0 |
| 4 | scaffold156626.9213 | 6 | A | G | 0.500 0 | 0.500 0 |
| 5 | Gene.280112.80 | 12 | C | T | 0.500 0 | 0.500 0 |
| 6 | super3477.582311 | 3 | G | A | 0.486 8 | 0.499 7 |
| 7 | super3658.67227 | 12 | G | C | 0.486 8 | 0.499 7 |
| 8 | super3673.196162 | 1 | G | C | 0.486 8 | 0.499 7 |
| 9 | scaffold4919.104166 | 11 | C | T | 0.486 8 | 0.499 7 |
| 10 | scaffold8912.56207 | 2 | C | T | 0.486 8 | 0.499 7 |
| 11 | scaffold18745.243173 | 7 | G | C | 0.486 8 | 0.499 7 |
| 12 | scaffold28210.166698 | 4 | C | T | 0.486 8 | 0.499 7 |
| 13 | scaffold45863.102243 | 8 | A | C | 0.486 8 | 0.499 7 |
| 14 | scaffold71374.123891 | 1 | G | T | 0.486 8 | 0.499 7 |
| 15 | scaffold120395.63994 | 12 | C | A | 0.486 8 | 0.499 7 |
| 16 | scaffold127392.228550 | 9 | A | C | 0.486 8 | 0.499 7 |
| 17 | scaffold144338.102562 | 10 | A | G | 0.486 8 | 0.499 7 |
| 18 | scaffold146854.112944 | 5 | A | T | 0.486 8 | 0.499 7 |
| 19 | Gene.117474.537 | 7 | T | C | 0.486 8 | 0.499 7 |
| 20 | super2715.43005 | 9 | C | T | 0.460 5 | 0.496 9 |

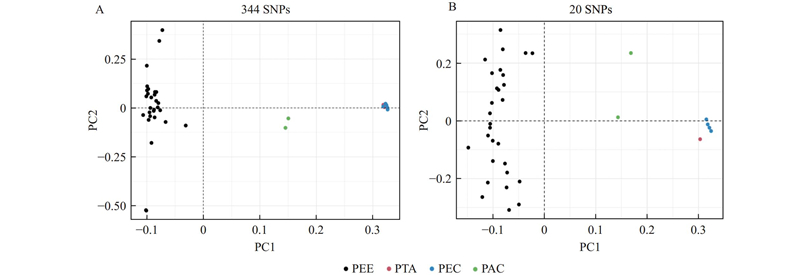
图3
基于344个SNPs(A)和20个SNPs(B)的主成分分析 黑色圆圈代表湿地松(PEE)样本,红色圆圈代表火炬松(PTA)样本,绿色圆圈代表湿加松(PEC)样本,蓝色圆圈代表火加松(PAC)样本。The black circle represents Pinus elliottii (PEE) samples. The red circle represents P. taeda (PTA) samples. The green circle represents P. elliottii × P. caribaea (PEC) samples. The blue circle represents P. taeda × P. caribaea (PAC) samples."
| 丁显印, 陶学雨, 刁 姝, 等. 2020. Pilodyn和Resistograph对湿地松活立木基本密度的评估. 南京林业大学学报(自然科学版), 44(3): 142−148. | |
| Ding X Y, Tao X Y, Diao S, et al. Estimation of wood basic density in a Pinus elliottii stand using Pilodyn and Resistograph measurements, Journal of Nanjing Forestry University (Natural Sciences Edition), 44(3): 142−148. [in Chinese] | |
|
樊晓静, 于文涛, 蔡春平, 等. 利用SNP标记构建茶树品种资源分子身份证. 中国农业科学, 2021, 54 (8): 1751- 1772.
doi: 10.3864/j.issn.0578-1752.2021.08.014 |
|
|
Fan X J, Yu W T, Cai C P, et al. Construction of molecular ID for tea cultivars by using of single-nucleotide polymorphism (SNP) markers. Scientia Agricultura Sinica, 2021, 54 (8): 1751- 1772.
doi: 10.3864/j.issn.0578-1752.2021.08.014 |
|
|
林 萍, 王开良, 姚小华, 等. 基于转录组SNP构建油茶主要品种资源的分子身份证. 中国农业科学, 2023, 56 (2): 217- 235.
doi: 10.3864/j.issn.0578-1752.2023.02.002 |
|
|
Lin P, Wang K L, Yao X H, et al. Development of DNA molecular ID in Camellia oleifera germplasm based on transcriptome-wide SNPs. Scientia Agricultura Sinica, 2023, 56 (2): 217- 235.
doi: 10.3864/j.issn.0578-1752.2023.02.002 |
|
| 刘超凡. 2022. 杨树无性系SSR指纹图谱及核心种质库构建. 长沙: 中南林业科技大学. | |
| Liu C F. 2022. Construction of SSR fingerprint database and core collection of Populus clones. Changsha: Central South University of Forestry & Technology. [in Chinese] | |
| 鲁仪增. 2019. 国槐遗传多样性评价及无性系分子鉴别. 北京: 中国林业科学研究院. | |
| Lu Y Z, 2019. Genetic diversity of Sophora japonica Linn. and clonal identification using molecular markers. Beijing: Chinese Academy of Forestry. [in Chinese] | |
|
栾启福, 姜景民, 张建忠, 等. 火炬松×加勒比松F_1代生长、树干通直度和基本密度遗传和配合力分析. 林业科学, 2011, 47 (3): 178- 183.
doi: 10.11707/j.1001-7488.20110327 |
|
|
Luan Q F, Jiang J M, Zhang J Z, et al. Estimation of heritability and combining ability for growth, stem-straightness and wood density of the F_1 generation of Pinus taeda × P. caribaea. Scientia Silvae Sinicae, 2011, 47 (3): 178- 183.
doi: 10.11707/j.1001-7488.20110327 |
|
| 栾启福, 李彦杰, 姜景民. 2014. 国外松种间杂交后代生长和形态性状变异及相关性分析. 植物研究, 34(1): 95–102. | |
| Luan Q F, Li Y J, Jiang J M, 2014. Variation of stem growth and morphology traits of exotic pine hybrids and their correlations. Bulletin of Botanical Research, 34(1): 95–102. [in Chinese] | |
| 栾启福. 2010. 几种松树杂交选育及其遗传分析. 北京: 中国林业科学研究院. | |
| Luan Q F. 2010. Hybridization of several pines and their genetical analysis. Beijing: Chinese Academy of Forestry. [in Chinese] | |
|
马庆国, 宋晓波, 贺君星, 等. 基于SSR分子标记的核桃种质资源分子身份证构建. 植物资源与环境学报, 2023, 32 (2): 1- 9.
doi: 10.3969/j.issn.1674-7895.2023.02.01 |
|
|
Ma Q G, Song X B, He J X, et al. Establishment of molecular identity cards of walnut (Juglans spp.) germplasm resources based on SSR molecular marker. Journal of Plant Resources and Environment, 2023, 32 (2): 1- 9.
doi: 10.3969/j.issn.1674-7895.2023.02.01 |
|
| 申 展. 2017. 基于形态学特性和SSR分子标记的文冠果种质资源评价与选优研究. 北京: 北京林业大学. | |
| Shen Z. 2017. Valuation and selection of Xanthoceras sorbifolium Bunge germplasm by morphology characteristics and SSR markers. Beijing: Beijing Forestry University. [in Chinese] | |
| 张良波. 2020. 光皮树优树群体遗传变异分析及核心种质构建. 北京: 北京林业大学. | |
| Zhang L B. 2020. Analysis of population genetic variation of plus trees and construction of core collection in Cornus wilsoniana. Beijing: Beijing Forestry University. [in Chinese] | |
| Batley J, Edwards D. 2007, SNP applications in plants//Oraguzie N C, Rikkerink E H A, Gardiner S E. eds. Association mapping in plants. New York, NY: Springer, 95−102. | |
|
Boban S, Maurya S, Jha Z. DNA fingerprinting: an overview on genetic diversity studies in the botanical taxa of Indian bamboo. Genetic Resource Crop Evolution, 2022, 69 (2): 469- 498.
doi: 10.1007/s10722-021-01280-8 |
|
|
Borders B E, Bailey R L. Loblolly pine—pushing the limits of growth. Southern Journal of Applied Forestry, 2001, 25 (2): 69- 74.
doi: 10.1093/sjaf/25.2.69 |
|
| Carvalho A, Matos M, Lima-Brito J, et al. DNA fingerprint of F1 interspecific hybrids from the Triticeae tribe using ISSRs. Euphytica, 2005, 143 (1/2): 93- 99. | |
|
Chambers G K, Curtis C, Millar C D, et al. DNA fingerprinting in zoology: past, present, future. Investigative Genetics, 2014, 5 (1): 3.
doi: 10.1186/2041-2223-5-3 |
|
|
Dallas J F. Detection of DNA “fingerprints” of cultivated rice by hybridization with a human minisatellite DNA probe. Proceedings of the National Academy of Sciences of the United States of America, 1988, 85 (18): 6831- 6835.
doi: 10.1073/pnas.85.18.6831 |
|
|
DeLaTorre, A R, Birol I, Bousquet J, et al. Insights into conifer Giga-genomes. Plant Physiology, 2014, 166 (4): 1724- 1732.
doi: 10.1104/pp.114.248708 |
|
|
DeVerno, L L, Mosseler A. Genetic variation in red pine (Pinus resinosa) revealed by RAPD and RAPD-RFLP analysis. Canadian Journal of Forest Research, 1997, 27 (8): 1316- 1320.
doi: 10.1139/x97-090 |
|
|
Diao S, Ding X Y, Luan Q F, et al. Development of 51 K liquid-phased probe array for Loblolly and Slash pines and its application to GWAS of Slash pine breeding population. Industrial Crops and Products, 2024, 216, 118777.
doi: 10.1016/j.indcrop.2024.118777 |
|
|
Ding X Y, Li Y J, Zhang Y N, et al. Genetic analysis and elite tree selection of the main resin components of Slash pine. Frontier Plant Science, 2023, 14, 1079952.
doi: 10.3389/fpls.2023.1079952 |
|
|
Duan Y F, Liu J, Xu J F, et al. DNA fingerprinting and genetic diversity analysis with simple sequence repeat markers of 217 potato cultivars (Solanum tuberosum L.) in China. American Journal of Potato Research, 2019, 96 (1): 21- 32.
doi: 10.1007/s12230-018-9685-6 |
|
| Ganal M W, Wieseke R, Luerssen H, et al. 2014. High-throughput SNP profiling of genetic resources in crop plants using genotyping arrays//Tuberosa R, Graner A, Frison E. eds. Genomics of plant genetic resources: volume 1. managing, sequencing and mining genetic resources. Dordrecht: Springer Netherlands, 113–130. | |
|
Ganea S, Ranade S S, Hall D, et al. Development and transferability of two multiplexes nSSR in Scots pine (Pinus sylvestris L.). Journal of Forestry Research, 2015, 26 (2): 361- 368.
doi: 10.1007/s11676-015-0042-z |
|
|
Geraldes A, DiFazio S P, Slavov G T, et al. A 34K SNP genotyping array for Populus trichocarpa: design, application to the study of natural populations and transferability to other Populus species. Molecular Ecology Resources, 2013, 13 (2): 306- 323.
doi: 10.1111/1755-0998.12056 |
|
|
Graham N, Telfer E, Frickey T, et al. Development and validation of a 36K SNP array for radiata pine (Pinus radiata D. Don). Forests, 2022, 13 (2): 176.
doi: 10.3390/f13020176 |
|
|
Guo Z F, Yang Q, Huang F F, et al. Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip. Plant Communications, 2021, 2 (6): 100230.
doi: 10.1016/j.xplc.2021.100230 |
|
|
Jackson C, Christie N, Reynolds S M, et al. A genome-wide SNP genotyping resource for tropical pine tree species. Molecular Ecology Resources, 2022, 22 (2): 695- 710.
doi: 10.1111/1755-0998.13484 |
|
| Jeffreys A J, Pena S D J. 1993. Brief introduction to human DNA fingerprinting//Pena S D J, Chakraborty R, Epplen J T, et al. eds. DNA fingerprinting: state of the science. Basel: Birkhäuser Verlag, 1–20. | |
|
Jeffreys A J, Wilson V, Thein S L. Hypervariable ‘minisatellite’ regions in human DNA. Nature, 1985, 314 (6006): 67- 73.
doi: 10.1038/314067a0 |
|
| Jehan T, Lakhanpaul S. Single nucleotide polymorphism (SNP)-methods and applications in plant genetics: a review. Indian Journal of Biotechnology, 2006, 5 (4): 435- 459. | |
| Jiang G L. 2015. Molecular marker-assisted breeding: a plant breeder’s review//Al-Khayri J M, Jain S M, Johnson D V. eds. Advances in plant breeding strategies: breeding, biotechnology and molecular tools. Cham: Springer International Publishing, 431–472. | |
| Kumar P, Gupta V K, Misra A K, et al. Potential of molecular markers in plant biotechnology. Plant Omics, 2020, 2 (4): 141- 162. | |
|
Lai M, Dong L M, Su R F, et al. Needle functional features in contrasting yield phenotypes of slash pine at three locations in southern China. Industrial Crops and Products, 2023, 206, 117613.
doi: 10.1016/j.indcrop.2023.117613 |
|
| Mammadov J, Aggarwal R, Buyyarapu R, et al. SNP markers and their impact on plant breeding. International Journal of Plant Genomics, 2012, 2012, 728398. | |
| Montanari S, Deng C, Koot E, et al. A multiplexed plant–animal SNP array for selective breeding and species conservation applications. G3 Genes| Genomes| Genetics, 2023, 13 (10): jkad170. | |
| Nadeem M A, Nawaz M A, Shahid M Q, et al. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnology & Biotechnological Equipment, 2018, 32 (2): 261- 285. | |
|
Nesbitt K A, Potts B M, Vaillancourt R E, et al. Partitioning and distribution of RAPD variation in a forest tree species, Eucalyptus globulus (Myrtaceae). Heredity, 1995, 74 (6): 628- 637.
doi: 10.1038/hdy.1995.86 |
|
|
Noormohammadi Z, Ibrahim-Khalili N, Sheidai M, et al. Genetic fingerprinting of diploid and tetraploid cotton cultivars by retrotransposon-based markers. Nucleus, 2018, 61 (2): 137- 143.
doi: 10.1007/s13237-018-0237-8 |
|
|
Nybom H, Weising K, Rotter B. DNA fingerprinting in botany: past, present, future. Investigative Genetics, 2014, 5 (1): 1.
doi: 10.1186/2041-2223-5-1 |
|
| Pavy N, Gagnon F, Rigault P, et al. Development of high-density SNP genotyping arrays for white spruce (Picea glauca) and transferability to subtropical and nordic congeners. Molecular Ecology Resources, 2013, 13 (2): 324- 336. | |
|
Prunier J, Verta J P, MacKay J J. Conifer genomics and adaptation: at the crossroads of genetic diversity and genome function. New Phytologist, 2016, 209 (1): 44- 62.
doi: 10.1111/nph.13565 |
|
| Rawat A, Barthwal S, Ginwal H S. Comparative assessment of SSR, ISSR and AFLP markers for characterization of selected genotypes of Himalayan Chir pine (Pinus roxburghii Sarg.) based on resin yield. Silvae Genetica, 2014, 63 (1): 94- 108. | |
|
Rodríguez S M, Ordás R, Alvarez J M. Conifer biotechnology: an overview. Forests, 2022, 13 (7): 1061.
doi: 10.3390/f13071061 |
|
|
Sousa-Santos C, Robalo J I, Collares-Pereira M J, et al. Heterozygous indels as useful tools in the reconstruction of DNA sequences and in the assessment of ploidy level and genomic constitution of hybrid organisms: Short Communication. DNA Sequence, 2005, 16 (6): 462- 467.
doi: 10.1080/10425170500356065 |
|
|
Tsumura Y, Yoshimura K, Tomaru N, et al. Molecular phytogeny of conifers using RFLP analysis of PCR-amplified specific chloroplast genes. Theoretical and Applied Genetics, 1995, 91 (8): 1222- 1236.
doi: 10.1007/BF00220933 |
|
|
Wang X R, Szmidt A E. Molecular markers in population genetics of forest trees. Scandinavian Journal of Forest Research, 2001, 16 (3): 199- 220.
doi: 10.1080/02827580118146 |
|
|
Wang Y Y, Lü H K, Xiang X H, et al. Construction of a SNP fingerprinting database and population genetic analysis of cigar tobacco germplasm resources in China. Frontiers in Plant Science, 2021, 12, 618133.
doi: 10.3389/fpls.2021.618133 |
|
|
Weiss-Schneeweiss H, Tremetsberger K, Schneeweiss G M, et al. Karyotype diversification and evolution in diploid and polyploid South American Hypochaeris (Asteraceae) inferred from rDNA localization and genetic fingerprint data. Annals of Botany, 2008, 101 (7): 909- 918.
doi: 10.1093/aob/mcn023 |
|
|
Wingett S W, Andrews S. FastQ screen: a tool for multi-genome mapping and quality control. F1000Research, 2018, 7, 1338.
doi: 10.12688/f1000research.15931.1 |
|
|
Wünsch A, Hormaza J I. Cultivar identification and genetic fingerprinting of temperate fruit tree species using DNA markers. Euphytica, 2002, 125 (1): 59- 67.
doi: 10.1023/A:1015723805293 |
|
|
Yang Y Y, Lyu M, Liu J, et al. Construction of an SNP fingerprinting database and population genetic analysis of 329 cauliflower cultivars. BMC Plant Biology, 2022, 22 (1): 522.
doi: 10.1186/s12870-022-03920-2 |
|
|
Zhang Y, Luan Q F, Jiang J J, et al. Prediction and utilization of malondialdehyde in exotic pine under drought stress using near-infrared spectroscopy. Frontiers in Plant Science, 2021, 12, 735275.
doi: 10.3389/fpls.2021.735275 |
|
|
Zhao D H, Bullock B P, Montes C R, et al. Loblolly pine outperforms slash pine in the southeastern United States – a long-term experimental comparison study. Forest Ecology and Management, 2019, 450, 117532.
doi: 10.1016/j.foreco.2019.117532 |
|
|
Zimin A, Stevens K A, Crepeau M W, et al. Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics, 2014, 196 (3): 875- 890.
doi: 10.1534/genetics.113.159715 |
| [1] | 李威, 于珍珍, 何徽, 赵佳璐, 刘西军. 碳输入改变对湿地松人工成熟林土壤微生物群落的影响[J]. 林业科学, 2025, 61(6): 232-242. |
| [2] | 李丰钰,黄平,郑勇奇,李长红,张雨婷,薛克娜,宗亦臣,赵鸿杰. 基于SSR标记的山茶属位点组合品种鉴别能力分析评价[J]. 林业科学, 2023, 59(8): 74-84. |
| [3] | 李彤彤,郭素娟,李艳华. 基于叶片形态数字化分析的板栗品种鉴别[J]. 林业科学, 2023, 59(3): 115-126. |
| [4] | 王哲,刘阳,赵奋成,曾明,李福明,吴惠姗,李义良,廖仿炎,邓乐平,钟岁英,郭文冰. 不同促脂剂对湿地松家系的增脂效果及对生长的影响[J]. 林业科学, 2022, 58(9): 106-116. |
| [5] | 刘林,张旭,余素君,孙洪刚,姜景民,王宇华. 湿地松材脂兼用林最优轮伐期的经济分析——以江西省景德镇市枫树山林场为例[J]. 林业科学, 2022, 58(4): 62-73. |
| [6] | 房焕英,肖胜生,余小芳,熊永,欧阳勋志,秦晓蕾. 湿地松人工林土壤呼吸及其组分对模拟酸雨的响应[J]. 林业科学, 2021, 57(7): 20-31. |
| [7] | 叶琳峰,李彦,王忠媛,陆世通,潘天天,陈森,谢江波. 湿润地区3种松属植物枝和根导水系统的效率-安全关系[J]. 林业科学, 2021, 57(7): 194-204. |
| [8] | 徐有明,周彩霞,林汉,陶吉云,张菊花. 湿地松树木形成层恢复活动期、活动期和休眠期原始细胞超微结构变化[J]. 林业科学, 2020, 56(10): 145-153. |
| [9] | 赵奋成, 郭文冰, 钟岁英, 邓乐平, 吴惠姗, 林昌明, 廖仿炎, 谭志强, 李义良. 基于针刺仪测定技术的湿地松木材密度间接选择效果[J]. 林业科学, 2018, 54(10): 172-179. |
| [10] | 张帅楠, 栾启福, 姜景民. 基于无损检测技术的湿地松生长及材性性状遗传变异分析[J]. 林业科学, 2017, 53(6): 30-36. |
| [11] | 杨会肖, 刘天颐, 刘纯鑫, 王金榜, 黄少伟. 火炬松基因资源林的空间分析[J]. 林业科学, 2015, 51(11): 50-59. |
| [12] | 刘天颐, 杨会肖, 刘纯鑫, 王金榜, 黄少伟. 火炬松基因资源的育种值预测与选择[J]. 林业科学, 2014, 50(8): 60-67. |
| [13] | 刘天颐;刘纯鑫;黄少伟;钟伟华;陈炳铨. 火炬松核心育种群体子代生长变异与选择[J]. , 2013, 49(2): 27-32. |
| [14] | 杨予静;李昌晓;张晔;崔云风. 水淹-干旱交替胁迫对湿地松幼苗盆栽土壤营养元素含量的影响[J]. , 2013, 49(2): 61-71. |
| [15] | 刘亚梅;刘盛全. 人工倾斜火炬松3年生苗木应压木的解剖性质[J]. 林业科学, 2012, 48(1): 131-137. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||