

林业科学 ›› 2025, Vol. 61 ›› Issue (4): 140-152.doi: 10.11707/j.1001-7488.LYKX20240069
收稿日期:2024-02-01
出版日期:2025-04-25
发布日期:2025-04-21
通讯作者:
郭小勤
E-mail:xqguo@zafu.edu.cn
基金资助:
Xiaoling Yan,Qin Hao,Zi Shen,Yujia Zhang,Xiaoqin Guo*( )
)
Received:2024-02-01
Online:2025-04-25
Published:2025-04-21
Contact:
Xiaoqin Guo
E-mail:xqguo@zafu.edu.cn
摘要:
目的: 分析毛竹PheFT1基因的表达、蛋白互作及生物学功能,为揭示该基因调控开花机理提供理论依据。方法: 利用实时荧光定量PCR检测PheFT1的组织特异性表达及对光周期的应答反应;采用PEG介导法分析PheFT1蛋白定位;应用农杆菌介导法将PheFT1基因转入拟南芥获得过表达植株及ft突变体回补植株,比较过表达植株与野生型植株、ft突变体回补植株与ft突变体植株之间的表型差异分析其生物学功能;基于酵母双杂交和双分子荧光互补试验分析PheFT1的互作蛋白。结果: 生物信息学分析结果显示,PheFT1基因CDS全长537 bp,编码178个氨基酸,属PEBP蛋白家族。亚细胞定位结果显示,PheFT1蛋白定位于细胞核和细胞质。实时荧光定量PCR结果显示,PheFT1基因在根、茎、叶和侧芽中均有表达,在侧芽和茎中表达量较高,在叶中表达量最低;长日照下PheFT1有较强的昼夜节律。异位表达结果显示,PheFT1基因会使拟南芥开花提前、茎变细和株高变矮。蛋白互作结果显示,PheFT1可与PheGF14和PheFD蛋白互作。结论: 毛竹PheFT1不仅是开花促进因子,还参与茎秆发育和高生长,该研究为进一步揭示PheFT1基因参与毛竹开花的分子机制及生长发育提供了参考。
中图分类号:
闫小玲,郝琴,申孜,张雨佳,郭小勤. 毛竹PheFT1基因的表达、蛋白互作及生物学功能分析[J]. 林业科学, 2025, 61(4): 140-152.
Xiaoling Yan,Qin Hao,Zi Shen,Yujia Zhang,Xiaoqin Guo. Expression, Protein Interaction and Biological Function Analysis of PheFT1 Gene in Moso Bamboo[J]. Scientia Silvae Sinicae, 2025, 61(4): 140-152.
表1
本研究中使用的 PCR 引物"
| 引物名称 Primer name | 引物序列(5'–3') Primer sequence (5'–3') | 引物用途 Application |
| PheFT1-F | ATGGCCAGCGGCAGCGTG | 基因克隆 |
| PheFT1-R | TCACATTCTTCTCCCACCAGTTCCCGA | Gene cloning |
| PheFT1-pCAMBIA1300-F | GAGCTCGGTACCCGGGGATCCATGGCCAGCGGCAGCGTG | 构建亚细胞定位载体 |
| PheFT1-pCAMBIA1300-R | CATGTCGACTCTAGAGGATCCCATTCTTCTCCCACCAGTTCCCGA | Construction of subcellular localization plasmid ppplasmidplasmids |
| PheFT1-QF | GGTGATCGGCGACGTGGT | |
| PheFT1-QR | TGTAGAATGTGCGCATGTCC | 荧光定量 PCR |
| PheTIP41-QF | AAAATCATTGTAGGCCATTGTCG | Quantitative real time PCR |
| PheTIP41-QR | ACTAAATTAAGCCAGCGGGAGTG | |
| PheFT1-pCAMBIA1300s-F | GGTACCCGGGGATCCTCTAGAATGGCCAGCGGCAGCGTG | 构建过表达载体 |
| PheFT1-pCAMBIA1300s-R | CTTGCATGCCTGCAGGTCGACTCACATTCTTCTCCCACCAGTTCCCGA | Construction of overexpression plasmid |
| PheFT1-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGGCCAGCGGCAGCGTG | |
| PheFT1-pGADT7-R | GCCGACATGTTTTTTCCCGGG TCACATTCTTCTCCCACCAGTTCCCGA | |
| PheFT1-pGBKT7-F | GGCCGAATTCCCGGGGATGGCCAGCGGCAGCGTG | |
| PheFT1-pGBKT7-R | CCGCTGCAGGTCGACGGATCC TCACATTCTTCTCCCACCAGTTCCCGA | |
| PheGF14a-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGGGTACGGCAGCGGGA | |
| PheGF14a-pGADT7-R | GCCGACATGTTTTTTCCCGGGCTAGTGCTCATCCTCGGGCTT | |
| PheGF14b-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGTCGGCACCTGCGGAG | |
| PheGF14b-pGADT7-R | GCCGACATGTTTTTTCCCGGGTTATCCAGAGCAAACAGC | |
| PheGF14c-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGTCGCGGGAGGAGAATG | |
| PheGF14c-pGADT7-R | GCCGACATGTTTTTTCCCGGGTCACTGGCCCTCGCCGGC | 构建酵母杂交载体 |
| PheGF14d-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGTCGCCGGCGGAGCCG | Construction of yeast two-hybrid plasmids |
| PheGF14d-pGADT7-R | GCCGACATGTTTTTTCCCGGGTCACAGTCCATCTCCGGATTCC | |
| PheGF14e-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGTCGCAGCCTGCCGAG | |
| PheGF14e-pGADT7-R | GCCGACATGTTTTTTCCCGGGTTACTGCCCATCTCCAGATTCG | |
| PheGF14f-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGTCTACTGCTGAAGCATCCCA | |
| PheGF14f-pGADT7-R | GCCGACATGTTTTTTCCCGGGTTAGTGCCCCTCTCCTTCAGG | |
| PheGF14g-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGGCGTCGCGCGAGGAT | |
| PheGF14g-pGADT7-R | GCCGACATGTTTTTTCCCGGGTCAACCAGATATACGAGCATCAGC | |
| PheGF14h-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGGAGGAGAGGGAGAAGGTCG | |
| PheGF14h-pGADT7-R | GCCGACATGTTTTTTCCCGGGCTAGAAACCTCCAGTTAGTTCTGACG | |
| PheFD-pGADT7-F | GAGTGGCCATTATGGCCCGGGATGGCCAACTACCACCACTA | |
| PheFD-pGADT7-R | GCCGACATGTTTTTTCCCGGGTCATCCTCTTAATTTTGTACTGCCA | |
| PheFT1-pSAT1-F | AGATCTCGAGCTCAAGCTTCGATGGCCAGCGGCAGCGTG | |
| PheFT1-pSAT1-R | GGTACCGTCGACTGCAGAATTTCACATTCTTCTCCCACCAGTTCCCGA | |
| PheFT1-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGGCCAGCGGCAGCGTG | |
| PheFT1-pSAT4-R | GGTACCGTCGACTGCAGAATTTCACATTCTTCTCCCACCAGTTCCCGA | |
| PheGF14a-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGGGTACGGCAGCGGGA | |
| PheGF14a-pSAT4-R | GGTACCGTCGACTGCAGAATTCTAGTGCTCATCCTCGGGCTT | |
| PheGF14b-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGTCGGCACCTGCGGAG | |
| PheGF14b-pSAT4-R | GGTACCGTCGACTGCAGAATTTTATCCAGAGCAAACAGC | |
| PheGF14c-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGTCGCGGGAGGAGAATG | |
| PheGF14c-pSAT4-R | GGTACCGTCGACTGCAGAATTTCACTGGCCCTCGCCGGC | 构建BiFC载体 |
| PheGF14d-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGTCGCCGGCGGAGCCG | Construction of BiFC plasmids |
| PheGF14d-pSAT4-R | GGTACCGTCGACTGCAGAATTTCACAGTCCATCTCCGGATTCC | |
| PheGF14e-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGTCGCAGCCTGCCGAG | |
| PheGF14e-pSAT4-R | GGTACCGTCGACTGCAGAATTTTACTGCCCATCTCCAGATTCG | |
| PheGF14f-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGTCTACTGCTGAAGCATCCCA | |
| PheGF14f-pSAT4-R | GGTACCGTCGACTGCAGAATTTTAGTGCCCCTCTCCTTCAGG | |
| PheGF14g-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGGCGTCGCGCGAGGAT | |
| PheGF14g-pSAT4-R | GGTACCGTCGACTGCAGAATTTCAACCAGATATACGAGCATCAGC | |
| PheGF14h-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGGAGGAGAGGGAGAAGGTCG | |
| PheGF14h-pSAT4-R | GGTACCGTCGACTGCAGAATTCTAGAAACCTCCAGTTAGTTCTGACG | |
| PheFD-pSAT4-F | AGATCTCGAGCTCAAGCTTCGATGGCCAACTACCACCACTA | |
| PheFD-pSAT4-R | GGTACCGTCGACTGCAGAATTTCATCCTCTTAATTTTGTACTGCCA |
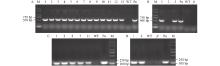
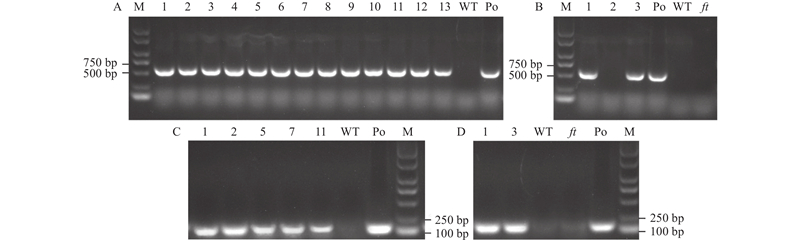
图4
过表达PheFT1植株及ft突变体回补植株PCR鉴定 A:过表达PheFT1植株的DNA鉴定结果。1–13:过表达PheFT1的植株;WT:野生型植株;Po:阳性对照(含35S::PheFT1的质粒);M:DL 2 000 plus DNA标记;B:PheFT1回补ft突变体植株的DNA鉴定结果。1–3:PheFT1回补ft突变体植株;Po:阳性对照(含35S::PheFT1的质粒);WT:野生型植株;ft:ft突变体植株;M:DL 2 000 plus DNA标记。C:过表达PheFT1植株的RNA鉴定结果。1、2、5、7和11:过表达PheFT1的植株;WT:野生型植株;Po:阳性对照(含35S::PheFT1的质粒);M:DL 2 000 plus DNA标记;D:PheFT1回补ft突变体植株的RNA鉴定结果。1和3:PheFT1回补ft突变体植株;WT:野生型植株;ft:ft突变体植株;Po:阳性对照(含35S::PheFT1的质粒);M:DL 2000 plus DNA标记。"
|
李建华, 岳晋军, 李海涛. 毛竹林经济和生态公益价值综合评价. 现代园艺, 2012, 18, 6- 7.
doi: 10.3969/j.issn.1006-4958.2012.01.003 |
|
|
Li J H, Yue J J, Li H T. Evaluation of economic and ecosystem services of moso bamboo stands. Contemporary Horticulture, 2012, 18, 6- 7.
doi: 10.3969/j.issn.1006-4958.2012.01.003 |
|
| 刘 丽, 陈骄羽, 邵明侠, 等. 毛竹PheFT6和PheFT17基因对外界环境的应答及蛋白互作分析. 农业生物技术学报, 2021, 29 (3): 506- 520. | |
| Liu L, Chen J Y, Shao M X, et al. Responses of PheFT6 and PheFT17 genes in Phyllostachys pubescens to external environment and protein interaction analysis. Journal of Agricultural Biotechnology, 2021, 29 (3): 506- 520. | |
| 罗 维, 牟 琼, 舒健虹, 等. 高羊茅FaFT基因表达, 蛋白互作及生物学功能分析. 生物技术通报, 2021, 37 (4): 8- 17. | |
| Luo W, Mou Q, Shu J H, et al. Expression, protein interactions and biological function analysis of FaFT in Festuca arundinacea. Biotechology Bulletin, 2021, 37 (4): 8- 17. | |
|
苗雅慧, 鞠 丹, 梁珂豪, 等. 青杄转录因子基因PwNF-YB8的克隆与功能分析. 林业科学, 2021, 57 (5): 77- 92.
doi: 10.11707/j.1001-7488.20210508 |
|
|
Miao Y H, Ju D, Liang K H, et al. Cloning and functional analysis of transcription factor gene PwNF-YB8 from Picea wilsonii. Scientia Silvae Sinicae, 2021, 57 (5): 77- 92.
doi: 10.11707/j.1001-7488.20210508 |
|
|
张雨佳, 刘 丽, 邹龙海, 等. 毛竹PheFT12a基因过表达对拟南芥开花及芽发育的影响. 核农学报, 2023, 37 (11): 2142- 2150.
doi: 10.11869/j.issn.1000-8551.2023.11.2142 |
|
|
Zhang Y J, Liu L, Zou L H, et al. Effects of overexpression of the moso bamboo PheFT12a on flowering and bud development of Arabidopsis. Journal of Nuclear Agricultural Sciences, 2023, 37 (11): 2142- 2150.
doi: 10.11869/j.issn.1000-8551.2023.11.2142 |
|
|
Abe M, Kobayashi Y, Yamamoto S, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science, 2005, 309 (5737): 1052- 1056.
doi: 10.1126/science.1115983 |
|
|
André D, Marcon A, Lee K C, et al. FLOWERING LOCUS T paralogs control the annual growth cycle in Populus trees. Current Biology, 2022, 32 (13): 2988- 2996.
doi: 10.1016/j.cub.2022.05.023 |
|
|
Aung B, Gruber M Y, Amyot L, et al. MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnology Journal, 2015, 13 (6): 779- 790.
doi: 10.1111/pbi.12308 |
|
|
Böhlenius H, Huang T, Charbonnel-Campaa L, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science, 2006, 312 (5776): 1040- 1043.
doi: 10.1126/science.1126038 |
|
|
Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science, 2007, 316 (5827): 1030- 1033.
doi: 10.1126/science.1141752 |
|
|
Dutta S, Biswas P, Chakraborty S, et al. Identification, characterization and gene expression analyses of important flowering genes related to photoperiodic pathway in bamboo. BMC Genomics, 2018, 19 (1): 190.
doi: 10.1186/s12864-018-4571-7 |
|
|
Fan C J, Ma J M, Guo Q R, et al. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One, 2013, 8 (2): e56573.
doi: 10.1371/journal.pone.0056573 |
|
|
Fan C M, Hu R B, Zhang X M, et al. Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biology, 2014, 14 (1): 1- 14.
doi: 10.1186/1471-2229-14-1 |
|
|
Fan H J, Zhuo R Y, Wang H Y, et al. A comprehensive analysis of the floral transition in ma bamboo (Dendrocalamus latiflorus) reveals the roles of DlFTs involved in flowering. Tree Physiology, 2022, 42 (9): 1899- 1911.
doi: 10.1093/treephys/tpac035 |
|
| Fang M C, Zhou Z J, Zhou X S, et al. Overexpression of OsFTL10 induces early flowering and improves drought tolerance in Oryza sativa L. Peer J, 2019, 7 (2): e6422. | |
|
Fu J X, Yang L W, Dai S L. Identification and characterization of the CONSTANS-like gene family in the short-day plant Chrysanthemum lavandulifolium. Molecular Genetics and Genomics, 2015, 290 (3): 1039- 1054.
doi: 10.1007/s00438-014-0977-3 |
|
|
Gu H W, Zhang K M, Chen J, et al. OsFTL4, an FT-like gene, regulates flowering time and drought tolerance in rice (Oryza sativa L.). Rice, 2022, 15 (1): 1- 15.
doi: 10.1186/s12284-021-00548-y |
|
|
Guo D L, Li C, Dong R, et al. Molecular cloning and functional analysis of the FLOWERING LOCUS T (FT) homolog GhFT1 from Gossypium hirsutum. Journal of Integrative Plant Biology, 2015, 57 (6): 522- 533.
doi: 10.1111/jipb.12316 |
|
| Guo X Q, Wang Y, Wang Q, et al. 2016. Molecular characterization of FLOWERING LOCUS T (FT) genes from bamboo (Phyllostachys violascens). Journal of Plant Biochemistry & Biotechnology, 25(2): 168–178. | |
|
Hisamoto Y, Kobayashi M. Flowering habit of two bamboo species, Phyllostachys meyeri and Shibataea chinensis, analyzed with flowering gene expression. Plant Species Biology, 2013, 28 (2): 109- 117.
doi: 10.1111/j.1442-1984.2012.00369.x |
|
| Hou C J, Yang C H. Functional analysis of FT and TFL1 orthologs from orchid (Oncidium gower Ramsey) that regulate the vegetative to reproductive transition. Plant & Cell Physiology, 2009, 50 (8): 1544- 1557. | |
|
Imaizumi T, Schultz T F, Harmon F G, et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science, 2005, 309 (5732): 293- 297.
doi: 10.1126/science.1110586 |
|
| Izawa T, Oikawa T, Sugiyama N, et al. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes & Development, 2002, 16 (15): 2006- 2020. | |
|
Jeon J S, Lee S, Jung K H, et al. Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Molecular Breeding, 2000, 6 (6): 581- 592.
doi: 10.1023/A:1011388620872 |
|
|
Kardailsky I, Shukla V K, Ahn J H, et al. Activation tagging of the floral inducer FT. Science, 1999, 286 (5446): 1962- 1965.
doi: 10.1126/science.286.5446.1962 |
|
|
Kikuchi R, Kawahigashi H, Ando T, et al. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiology, 2009, 149 (3): 1341- 1353.
doi: 10.1104/pp.108.132134 |
|
|
Kinoshita T, Ono N, Hayashi Y, et al. FLOWERING LOCUS T regulates stomatal opening. Current Biology, 2011, 21 (14): 1232- 1238.
doi: 10.1016/j.cub.2011.06.025 |
|
|
Kobayashi Y, Kaya H, Goto K, et al. A pair of related genes with antagonistic roles in mediating flowering signals. Science, 1999, 286 (5446): 1960- 1962.
doi: 10.1126/science.286.5446.1960 |
|
| Kobayashi Y, Weigel D. Move on up, it's time for change: mobile signals controlling photoperiod-dependent flowering. Genes & Development, 2007, 21 (19): 2371- 2384. | |
|
Komiya R, Ikegami A, Tamaki S, et al. Hd3a and RFT1 are essential for flowering in rice. Development, 2008, 135 (4): 767- 774.
doi: 10.1242/dev.008631 |
|
|
Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development, 2009, 136 (20): 3443- 3450.
doi: 10.1242/dev.040170 |
|
|
Kong F J, Liu B H, Xia Z J, et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiology, 2010, 154 (3): 1220- 1231.
doi: 10.1104/pp.110.160796 |
|
|
Lee J H, Lee J S, Ahn J H. Ambient temperature signaling in plants: an emerging field in the regulation of flowering time. Journal of Plant Biology, 2008, 51 (5): 321- 326.
doi: 10.1007/BF03036133 |
|
|
Lee R, Baldwin S, Kenel F, et al. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nature Communications, 2013, 4, 2884.
doi: 10.1038/ncomms3884 |
|
|
Lin X C, Chow T Y, Chen H H, et al. Understanding bamboo flowering based on large-scale analysis of expressed sequence tags. Genetics and Molecular Research, 2010, 9 (2): 1085- 1093.
doi: 10.4238/vol9-2gmr804 |
|
|
Liu L, Liu C, Hou X L, et al. FTIP1 is an essential regulator required for florigen transport. PLoS Biology, 2012, 10 (4): e1001313.
doi: 10.1371/journal.pbio.1001313 |
|
|
Liu Y Y, Yang K Z, Wei X X, et al. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytologist, 2016, 212 (3): 730- 744.
doi: 10.1111/nph.14066 |
|
|
Lv Z Y, Zhang L, Chen L X, et al. The Artemisia annua FLOWERING LOCUS T Homolog 2, AaFT2, is a key regulator of flowering time. Plant Physiology and Biochemistry, 2018, 126, 197- 205.
doi: 10.1016/j.plaphy.2018.02.033 |
|
|
Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△Ct method. Methods, 2001, 25 (4): 402- 408.
doi: 10.1006/meth.2001.1262 |
|
|
Meng X, Muszynski M G, Danilevskay O N. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. The Plant Cell, 2011, 23 (3): 942- 960.
doi: 10.1105/tpc.110.081406 |
|
| Mouradov A, Cremer F, Coupland G. Control of flowering time: interacting pathways as a basis for diversity. The Plant Cell, 2002, 14 (Suppl): S111- S130. | |
| Nakanishi H, Nakamichi N, Ito S, et al. 2013. Clock-controlled and FLOWERING LOCUS T (FT)-dependent photoperiodic pathway in Lotus japonicus I: verification of the flowering-associated function of an FT homolog. Bioscience, Biotechnology and Biochemistry, 77(4): 747–753. | |
|
Patil H B, Chaurasia A K, Azeez A, et al. Characterization of two TERMINAL FLOWER1 homologs PgTFL1 and PgCENa from pomegranate (Punica granatum L.). Tree Physiology, 2018, 38 (5): 772- 784.
doi: 10.1093/treephys/tpx154 |
|
|
Peng Z H, Lu Y, Li L B, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nature Genetics, 2013, 45 (4): 456- 461.
doi: 10.1038/ng.2569 |
|
| Purwestri Y A, Ogaki Y, Tamaki S, et al. The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant & Cell Physiology, 2009, 50 (3): 429- 438. | |
|
Qin Z R, Wu J J, Geng S F, et al. Regulation of FT splicing by an endogenous cue in temperate grasses. Nature Communications, 2017, 8 (1): 14320.
doi: 10.1038/ncomms14320 |
|
|
Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences, 2011, 68 (12): 2013- 2037.
doi: 10.1007/s00018-011-0673-y |
|
|
Su Q, Chen L, Cai Y P, et al. Functional redundancy of FLOWERING LOCUS T 3b in soybean flowering time regulation. International Journal of Molecular Sciences, 2022, 23 (5): 2497.
doi: 10.3390/ijms23052497 |
|
|
Sun H B, Jia Z, Cao D, et al. GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS One, 2011, 6 (12): e29238.
doi: 10.1371/journal.pone.0029238 |
|
|
Takadas S, Goto K. TERMINAL FLOWER 2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN 1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. The Plant Cell, 2003, 15 (12): 2856- 2865.
doi: 10.1105/tpc.016345 |
|
|
Tamaki S, Matsuo S, Wong H L, et al. Hd3a protein is a mobile flowering signal in rice. Science, 2007, 316 (5827): 1033- 1036.
doi: 10.1126/science.1141753 |
|
| Teo C J, Takahashi K, Shimizu K, et al. Potato tuber induction is regulated by interactions between components of a tuberigen complex. Plant & Cell Physiology, 2017, 58 (2): 365- 374. | |
|
Triozzi P M, Ramos-Sánchez J M, Hernández-Verdeja T, et al. Photoperiodic regulation of shoot apical growth in poplar. Frontiers in Plant Science, 2018, 9, 1030.
doi: 10.3389/fpls.2018.01030 |
|
|
Wang F M, Yano K, Nagamatsu S, et al. Genome-wide expression quantitative trait locus studies facilitate isolation of causal genes controlling panicle structure. The Plant Journal, 2020, 103 (1): 266- 278.
doi: 10.1111/tpj.14726 |
|
| Wang H Y, Li J, Liu Z R, et al. 2022. Dwarf phenotype induced by overexpression of a GAI1-like gene from Rhus chinensis. Plant Cell, Tissue and Organ Culture, 151(3): 617–629. | |
|
Wigge P A, Kim M C, Jaeger K E, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science, 2005, 309 (5737): 1056- 1059.
doi: 10.1126/science.1114358 |
|
|
Woods D, Dong Y X, Bouche F, et al. A florigen paralog is required for short-day vernalization in a pooid grass. eLife, 2019, 8, e42153.
doi: 10.7554/eLife.42153 |
|
|
Wu J, Wu Q H, Bo Z J, et al. Comprehensive effects of Flowering Locus T-mediated stem growth in tobacco. Frontiers in Plant Science, 2022, 13, 922919.
doi: 10.3389/fpls.2022.922919 |
|
|
Yang Z Y, Chen L, Kohnen M V, et al. Identification and characterization of the PEBP family genes in moso bamboo (Phyllostachys heterocycla). Scientific Reports, 2019, 9 (1): 14998.
doi: 10.1038/s41598-019-51278-7 |
|
|
Zhan Z X, Zhang C S, et al. Molecular cloning, expression analysis, and subcellular localization of FLOWERING LOCUS T (FT) in carrot (Daucus carota L. ). Molecular Breeding, 2017, 37 (12): 149.
doi: 10.1007/s11032-017-0749-y |
|
|
Zhang H J, Zhang Y. Molecular cloning and functional characterization of CmFT (FLOWERING LOCUS T) from Cucumis melo L. Journal of Genetics, 2020, 99 (1): 1- 8.
doi: 10.1007/s12041-019-1160-8 |
|
|
Zhang J L, Chen L, Cai Y P, et al. A novel MORN-motif type gene GmMRF2 controls flowering time and plant height of soybean. International Journal of Biological Macromolecules, 2023, 245, 125464.
doi: 10.1016/j.ijbiomac.2023.125464 |
|
|
Zhang M D, Hu S, Yi F, et al. Organelle visualization with multicolored fluorescent markers in bamboo. Frontiers in Plant Science, 2021, 12, 658836.
doi: 10.3389/fpls.2021.658836 |
|
|
Zhao J W, Gao P J, Li C L, et al. PhePEBP family genes regulated by plant hormones and drought are associated with the activation of lateral buds and seedling growth in Phyllostachys edulis. Tree Physiology, 2019, 39 (8): 1387- 1404.
doi: 10.1093/treephys/tpz056 |
|
|
Zhu H G, Tian W G, Zhu X F, et al. Ectopic expression of GhSAMDC1 improved plant vegetative growth and early flowering through conversion of spermidine to spermine in tobacco. Scientific Reports, 2020, 10 (1): 14418.
doi: 10.1038/s41598-020-71405-z |
| [1] | 马欣欣,王游,王佳军,冯龙,马建锋. 热解过程中竹材灰分组成变化及硅的转化分布规律[J]. 林业科学, 2025, 61(2): 172-179. |
| [2] | 张翱,李文婷,王天祥,武耀星,雷刚,漆良华. 毛竹林土壤易氧化有机碳区域分异及影响因素[J]. 林业科学, 2024, 60(6): 1-12. |
| [3] | 朱永莉,张杰. 上海市栽培乐昌含笑开花及传粉生物学[J]. 林业科学, 2024, 60(6): 86-93. |
| [4] | 王一,栾军伟,陈琛,刘世荣. 毛竹林土壤呼吸及组分对氮磷添加的非对等响应[J]. 林业科学, 2023, 59(7): 54-64. |
| [5] | 袁金玲,岳晋军,马婧瑕,于磊,刘蕾. 元宝毛竹的秆形特征[J]. 林业科学, 2023, 59(5): 71-80. |
| [6] | 蔡宗明,邓智文,李秉钧,李士坤,温伟庆,荣俊冬,郑郁善,陈礼光. 带状采伐宽度对毛竹林地下竹鞭结构特征的影响[J]. 林业科学, 2023, 59(4): 79-87. |
| [7] | 郑亚雄,范少辉,张璇,周潇,官凤英. 带状采伐毛竹林生产力变化规律[J]. 林业科学, 2023, 59(2): 22-29. |
| [8] | 代琳心,王智辉,李振瑞,王佳军,刘杏娥,文甲龙,马建锋. 基于TG-FTIR的竹材细胞壁主要组分热解特性[J]. 林业科学, 2023, 59(11): 85-94. |
| [9] | 刘彩霞,陈俊辉,秦华,梁辰飞,徐秋芳. 有机无机肥长期配施对毛竹林土壤固碳和固氮微生物的影响[J]. 林业科学, 2022, 58(7): 82-92. |
| [10] | 马瑞乡,黄满昌,张佳佳,赵澳舜,丁兴萃,罗自生,刘胜辉,肖子璋,沈凯. 毛竹笋采后呼吸途径变化及高氧处理效应[J]. 林业科学, 2022, 58(6): 33-46. |
| [11] | 张玮,何玉友,郭子武,汪舍平,陈双林. 失管毛竹林演替过程中乔木树种群落结构和多样性特征[J]. 林业科学, 2022, 58(12): 12-20. |
| [12] | 李澍农,张亚梅,余养伦,于文吉. 水煮处理竹材的吸湿性和化学成分研究[J]. 林业科学, 2022, 58(1): 119-126. |
| [13] | 谢佳敏,周明兵. 毛竹Mariner-like element自主转座子的鉴定与生物信息学分析[J]. 林业科学, 2022, 58(1): 175-184. |
| [14] | 李真,袁婷婷,朱成磊,杨克彬,宋新章,高志民. 毛竹铵态氮转运蛋白的分子特征及基因表达模式[J]. 林业科学, 2021, 57(7): 70-79. |
| [15] | 袁雪,袁涛,刘少丹. 牡丹秋季开花过程中生理生化变化及DNA甲基化差异[J]. 林业科学, 2021, 57(5): 53-67. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||