

Scientia Silvae Sinicae ›› 2021, Vol. 57 ›› Issue (1): 40-52.doi: 10.11707/j.1001-7488.20210105
Previous Articles Next Articles
Hui Li1,2,Xinren Dai1,Zaizhi Zhou2,Quanzi Li1,*
Received:2020-03-16
Online:2021-01-01
Published:2021-03-10
Contact:
Quanzi Li
CLC Number:
Hui Li,Xinren Dai,Zaizhi Zhou,Quanzi Li. Screening and Co-Expression Network Building of Xylem Development-Related Genes in Larix kaempferi[J]. Scientia Silvae Sinicae, 2021, 57(1): 40-52.

Fig.4
WGCNA analysis for specifically expressed gene in xylem A: The process for screening soft threshold; B: The process for gene cluster and module classification; C: The correlation of genes in modules, light color means high correlation, and crineous means low correlation; D: The correlation of modules."


Fig.6
Expression abundance and validation for co-expression within the network A: The co-expression network for 17 hub genes(red circles), 11 cellulose and hemicelluloses formation-related genes(green triangles) and 14 lignin formation-related genes(blue rhombuses); B: Expression heatmap for genes within the network, the expression level of genes is standardized by FPKM; C: Validation of genes within network through AspWood(http://aspwood.popgenie.org)."
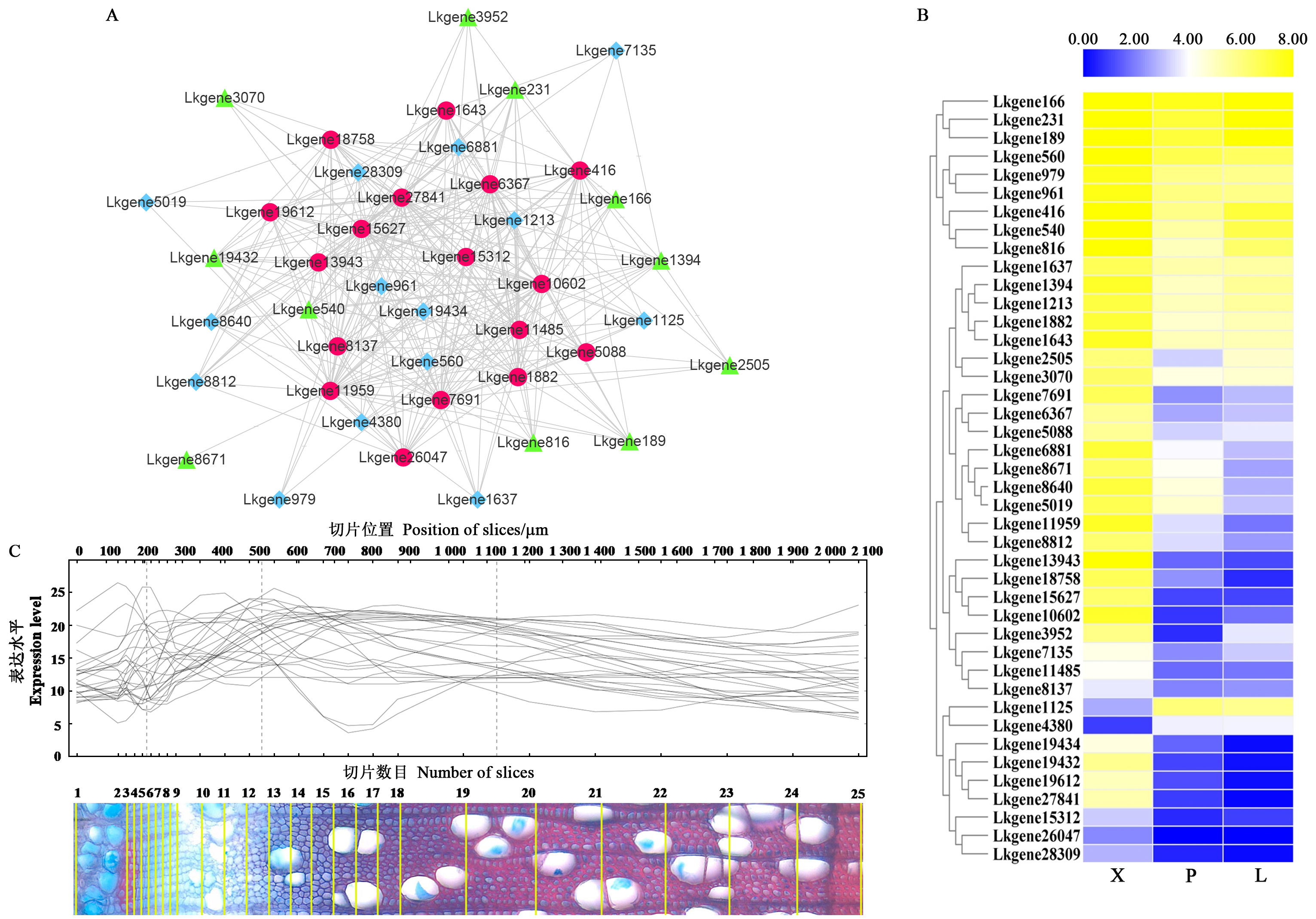
Table 2
Gene correspondence table between Larix kaempferi and Populus trichocarpa"
| 基因号 Gene ID | 毛果杨同源基因 Homolog in P.trichocarpa | 基因名 Gene name | 基因号 Gene ID | 毛果杨同源基因 Homolog in P.trichocarpa | 基因名 Gene name | |
| Lkgene 10602 | Potri.001G054600 | LAC17 | Lkgene8671 | Potri.005G195600 | PER12 | |
| Lkgene1213 | Potri.001G078900 | KOR | Lkgene6367 | Potri.005G203300 | F7D19.4 | |
| Lkgene5088 | Potri.001G083100 | MJC20.20 | Lkgene7691 | Potri.006G114400 | LRR-RLK | |
| Lkgene8137 | Potri.001G239300 | PLL4 | Lkgene11959 | Potri.006G136700 | SUS2 | |
| Lkgene19612 | Potri.001G312600 | UGT85A2 | Lkgene26047 | Potri.008G073700 | LAC12 | |
| Lkgene4380 | Potri.001G317600 | SPS3 | Lkgene13943 | Potri.010G141600 | HST | |
| Lkgene18758 Lkgene27841 | Potri.002G202300 | SUS2 | Lkgene1394 Lkgene2505 | Potri.003G183900 | HST | |
| Lkgene560 Lkgene979 Lkgene961 | Potri.004G089300 | F28G11.11 | Lkgene8640 Lkgene6881 Lkgene8812 Lkgene28309 Lkgene19434 | Potri.005G145300 | SBT1.7 | |
| Lkgene540 Lkgene816 | Potri.006G033300 | CYP98A3 | Lkgene166 Lkgene231 | Potri.013G157900 | CYP73A5 | |
| Lkgene416 | Potri.003G126800 | TUBB6 | Lkgene19432 | Potri.013G083600 | PER52 | |
| Lkgene1637 | Potri.003G151700 | KOR | Lkgene5019 | Potri.014G126900 | CEL1 | |
| Lkgene3952 | Potri.003G181400 | CCR1 | Lkgene1882 | Potri.014G197500 | — | |
| Lkgene3070 | Potri.015G003100 | OMTI | Lkgene7135 | Potri.017G029000 | T28P16.12 | |
| Lkgene1125 | Potri.017G057800 | SPS3 | Lkgene15312 | Potri.018G096100 | AXX17 | |
| Lkgene1643 | Potri.019G038200 | MS1 | Lkgene189 | Potri.019G049500 | 4CL2 | |
| Lkgene11485 | Potri.T070100.1 | MS2 | Lkgene15627 | — | — |
Table 3
Functional annotation for wood formation-related genes"
| 基因 Gene | 拟南芥同源基因 Homolog in Arabidopsis | 基因名 Gene name | 注释 Annotation | 参考文献 Reference |
| Lkgene393, 1210, 546, 10216, 1545, 189 | AT3G10340 | PAL4 | 催化L-苯丙氨酸生物合成中的第一反应Catalyze the first reaction in the biosynthesis from L-phenylalanine | |
| Lkgene3952 | AT1G15950 | CCR1 | 参与木质素合成的后期反应Involve in the latter stages of lignin biosynthesis | |
| Lkgene231, 166 | AT2G30490 | C4H | 调控合成木质素的碳代谢流Control carbon flux to lignins | |
| Lkgene19432 | AT5G05340 | PER52 | 木质素生物合成及降解Biosynthesis and degradation of lignin | |
| Lkgene4199, 8671 | AT1G71695 | PER12 | 木质素生物合成及降解Biosynthesis and degradation of lignin | |
| Lkgene1394, 2505 | AT5G48930 | HCT | 参与木质素合成Involve in the biosynthesis of lignin | |
| Lkgene3070, 834, 8363 | AT5G54160 | COMT1 | 催化木质素单体甲基化Catalyze the methylation of monolignols | |
| Lkgene5584, 540, 826, 816 | AT2G40890 | CYP98A3 | 参与木质素生物合成Essential for the biosynthesis of lignin | |
| Lkgene26047, 24638, 30550, 18632, 23499, 27977, 20214, 28123, 30125, 27556, 17746, 19212, 18598, 28597, 27567, 20222, 25143, 27539, 27532, 27443, 24826, 27783, 29817, 18090, 20304, 16114, 22264, 29559, 19527, 24209, 24522, 26047, 22691, 19315, 20120, 17571 | AT5G05390 | LAC12 | 木质素降解Degradation of lignin | |
| Lkgene2825, 12019, 19328, 27605, 21583, 13836, 13786, 5005, 16084, 12626, 17261, 16031, 5049, 5610, 15000, 10412, 25453, 30079, 29273, 20576, 10602, 13502, 15304 | AT5G60020 | LAC17 | 木质素单体聚合Polymerization of monolignols | |
| Lkgene24340, 114, 7969, 13943 | AT3G16920 | CTL2 | 纤维素合成及纤维素和半纤维素相互作用Biosynthesis of cellulose and interactions between hemicelluloses and cellulose | |
| Lkgene1782 | AT2G19860 | HXK2 | 调控细胞程序化死亡Regulate the execution of programmed cell death | |
| Lkgene2468, 1670, 3934, 811, 10000, 7280, 1786 | AT5G03760 | CSLA9 | 参与半乳甘露聚糖合成Participate in galactomannan biosynthesis | |
| Lkgene25783, 11296, 7496 | AT5G20950 | F22D1.120 | 参与木葡聚糖代谢Participate in xyloglucan metabolism | |
| Lkgene946, 1213, 1637 | AT5G49720 | DEC | 参与纤维素微纤维和次生细胞壁形成Require for cellulose microfibrils formation and secondary cell wall formation | |
| Lkgene5019 | AT1G70710 | CEL1 | 参与细胞壁纤维素形成Require for cellulose formation of the cell wall | |
| Lkgene1125, 5823, 4380, 6726, 12809, 2982 | AT1G04920 | SPS3 | 参与纤维伸长Regulate fiber elongation |
| 丁昌俊, 张伟溪, 高暝, 等. 不同生长势美洲黑杨转录组差异分析. 林业科学, 2016, 52 (3): 47- 58. | |
| Ding C J , Zhang W X , Gao M , et al. Analysis of transcriptome differences among Populus deltoides with different growth potentials. Scientia Silvae Sinicae, 2016, 52 (3): 47- 58. | |
|
孙晓梅, 张守攻, 李时元, 等. 日本落叶松纸浆材优良家系多性状联合选择. 林业科学, 2005, 41 (4): 48- 54.
doi: 10.3321/j.issn:1001-7488.2005.04.009 |
|
|
Sun X M , Zhang S G , Li S Y , et al. Multi-traits selection of open-pollinated Larix kaempferi families for pulpwood purpose. Scientia Silvae Sinicae, 2005, 41 (4): 48- 54.
doi: 10.3321/j.issn:1001-7488.2005.04.009 |
|
| 徐宗昌, 孔英珍. 普通烟草CESA基因家族成员的鉴定、亚细胞定位及表达分析. 遗传, 2017, 39 (6): 512- 524. | |
| Xu Z C , Kong Y Z . Genome-wide identification, subcellular location and gene expression analysis of the member of CEAS gene family in common tobacco(Nicotiana tabacum L. ). Herditas, 2017, 39 (6): 512- 524. | |
| 杨立. 2016. 杨树PtoVNS11和PtoMYB156转录因子在次生壁形成及类黄酮代谢途径中的功能分析. 重庆: 西南大学博士学位论文. | |
| Yang L. 2016. Functional analysis of PtoVNS11 and PtoMYB156 transcription factors involved in secondary wall formation and flavonoid biosynthetic pathway in poplar. Chongqing: PhD thesis of Southwest University.[in Chinese] | |
| 周贤武, 张俊珍, 周海宾, 等. 树龄对日本落叶松木材物理力学性质的影响. 林业工程学报, 2014, 28 (4): 54- 57. | |
| Zhou X W , Zhang J Z , Zhou H B , et al. Tree age's effects on physical and mechanical properties of Larix kaempferi wood. Journal of Forestry Engineering, 2014, 28 (4): 54- 57. | |
|
Allocco D J , Kohane I S , Butte A J . Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics, 2004, 5 (1): 18.
doi: 10.1186/1471-2105-5-18 |
|
|
Amann K , Lezhneva L , Wanner G , et al. ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of. Plant Cell, 2004, 16 (11): 3084- 3097.
doi: 10.1105/tpc.104.024935 |
|
|
Aoki K , Ogata Y , Shibata D . Approaches for extracting practical information from gene co-expression networks in plant biology. Plant and Cell Physiology, 2007, 48 (3): 381- 390.
doi: 10.1093/pcp/pcm013 |
|
|
Au K F , Underwood J G , Lee L , et al. Improving PacBio long read accuracy by short read alignment. PLoS ONE, 2012, 7 (10): e46679.
doi: 10.1371/journal.pone.0046679 |
|
|
Berthet S , Demont-Caulet N , Pollet B , et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell, 2011, 23 (3): 1124- 1137.
doi: 10.1105/tpc.110.082792 |
|
|
Brown D M , Zeef L A H , Ellis J , et al. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell, 2005, 17 (8): 2281- 2295.
doi: 10.1105/tpc.105.031542 |
|
| Carolin S , Bernard W , Soile J L , et al. Ethylene-related gene expression networks in wood formation. Frontiers in Plant Science, 2018, 272 (9): 1- 17. | |
|
Carruthers M , Yurchenko A A , Augley J J , et al. De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model Salmonid fish species. BMC Genomics, 2018, 19 (1): 32.
doi: 10.1186/s12864-017-4379-x |
|
|
Chano V , de Heredia U L , Collada C , et al. Transcriptomic analysis of juvenile wood formation during the growing season in Pinus canariensis. Holzforschung, 2017, 71 (12): 919- 937.
doi: 10.1515/hf-2017-0014 |
|
|
Chin C H , Chen S H , Wu H H , et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Systems Biology, 2014, (Suppl 4): S11.
doi: 10.1186/1752-0509-8-S4-S11 |
|
|
Conesa A , Götz S , García-Gómez J M , et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 2005, 21 (18): 3674- 3676.
doi: 10.1093/bioinformatics/bti610 |
|
|
Cronn R , Dolan P C , Jogdeo S , et al. Transcription through the eye of a needle: daily and annual cyclic gene expression variation in Douglas-fir needles. BMC Genomics, 2017, 18 (1): 558.
doi: 10.1186/s12864-017-3916-y |
|
|
Déjardin A , Laurans F , Arnaud D , et al. Wood formation in Angiosperms. Comptes Rendus Biologies, 2010, 333 (4): 325- 334.
doi: 10.1016/j.crvi.2010.01.010 |
|
|
Du H , Yu Y , Ma Y , et al. Sequencing and de novo assembly of a near complete indica rice genome. Nature Communications, 2017, 8, 15324.
doi: 10.1038/ncomms15324 |
|
|
Eid J , Fehr A , Gray J , et al. Real-time DNA sequencing from single polymerase molecules. Science, 2009, 323 (5910): 133- 138.
doi: 10.1126/science.1162986 |
|
|
Hertzberg M , Aspeborg H , Schrader J , et al. A transcriptional roadmap to wood formation. Proceedings of the National Academy of Sciences, 2001, 98, 14732- 14737.
doi: 10.1073/pnas.261293398 |
|
|
Jokipii-Lukkari S , Delhomme N , Schiffthaler B , et al. Transcriptional roadmap to seasonal variation in wood formation of Norway spruce. Plant Physiology, 2018, 176 (4): 2851- 2870.
doi: 10.1104/pp.17.01590 |
|
|
Kajita S , Hishiyama S , Tomimura Y , et al. Structural characterization of modified lignin in transgenic tobacco plants in which the activity of 4-coumarate: coenzyme A ligase is depressed. Plant Physiology, 1997, 114 (3): 871- 879.
doi: 10.1104/pp.114.3.871 |
|
| Kim D , Langmead B , Salzberg S L . HISAT: a fast spliced aligner with low memory requirements. Nature Methods, 2010, 12 (4): 357- 360. | |
|
Kim M , Lim J H , Ahn C S , et al. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell, 2006, 18 (9): 2341- 2355.
doi: 10.1105/tpc.106.041509 |
|
|
Kirst M , Johnson A F , Baucom C , et al. Apparent homology of expressed genes from wood-forming tissues of loblolly pine(Pinus taeda L. ) with Arabidopsis thaliana. Proceedings of the National Academy of Sciences, 2003, 100 (12): 7383- 7388.
doi: 10.1073/pnas.1132171100 |
|
|
Li J , Harata-Lee Y , Denton M D , et al. Long read reference genome-free reconstruction of a full-length transcriptome from Astragalus membranaceus reveals transcript variants involved in bioactive compound biosynthesis. Cell Discovery, 2017a, 3, 17031.
doi: 10.1038/celldisc.2017.31 |
|
|
Li W F , Yang W H , Zhang S G , et al. Transcriptome analysis provides insights into wood formation during larch tree aging. Tree Genetics & Genomes, 2017b, 13 (1): 1- 13.
doi: 10.1007/s11295-017-1106-3/fulltext.html |
|
|
Li W Z , Godzik A . Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 2006, 22 (13): 1658- 1659.
doi: 10.1093/bioinformatics/btl158 |
|
|
Lorenz W W , Yu Y S , Dean J F . An improved method of RNA isolation from loblolly pine(P. taeda L.) and other conifer species. Journal of Visualized Experiments, 2010, (36): 1751.
doi: 10.3791/1751 |
|
|
Lorenzo M , Pinedo M , Equiza M , et al. Changes in apoplastic peroxidase activity and cell wall composition are associated with cold-induced morpho-anatomical plasticity of wheat leaves. Plant Biology, 2019, (Suppl 1): 84- 94.
doi: 10.1111/plb.12709 |
|
|
Lukens L , Downs G . Bioinformatics techniques for understanding and analyzing tree gene expression data. Genomics of Tree Crops, 2012, 17- 38.
doi: 10.1007/978-1-4614-0920-5_2 |
|
|
Luo W , Cao X , Xu X , et al. Developmental transcriptome analysis and identification of genes involved in formation of intestinal air-breathing function of Dojo loach, Misgurnus anguillicaudatus. Scientific Reports, 2016, 6, 31845.
doi: 10.1038/srep31845 |
|
|
Lutfiyya L L , Xu N , D'Ordine R L , et al. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. Journal of Plant Physiology, 2007, 164 (7): 923- 933.
doi: 10.1016/j.jplph.2006.04.014 |
|
|
Metzker M L . Sequencing technologies-the next generation. Nature Reviews Genetics, 2010, 11 (1): 31- 46.
doi: 10.1038/nrg2626 |
|
|
Moreau C , Aksenov N , Lorenzo M G , et al. A genomic approach to investigate developmental cell death in woody tissues of Populus trees. Genome Biology, 2005, 6 (4): R34.
doi: 10.1186/gb-2005-6-4-r34 |
|
|
Moreau C , Pesquet E , Sjödin A , et al. A unique program for cell death in xylem fibers of Populus stem. The Plant Journal, 2009, 58 (2): 260- 274.
doi: 10.1111/j.1365-313X.2008.03777.x |
|
|
Nicol F , His I , Jauneau A , et al. A plasma membrane-bound putative endo-1, 4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. The EMBO Journal, 1998, 17 (19): 5563- 5576.
doi: 10.1093/emboj/17.19.5563 |
|
|
Ohashi-Ito K , Oda Y , Fukuda H . Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell, 2010, 22 (10): 3461- 3473.
doi: 10.1105/tpc.110.075036 |
|
|
Osakabe K , Tsao C C , Li L , et al. Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proceedings of the National Academy of Sciences, 1999, 96 (16): 8955- 8960.
doi: 10.1073/pnas.96.16.8955 |
|
|
Persson S , Wei H R , Milne J , et al. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proceedings of the National Academy of Sciences, 2005, 102 (24): 8633- 8638.
doi: 10.1073/pnas.0503392102 |
|
|
Prigge M J , Otsuga D , Alonso J M , et al. Class Ⅲ homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell, 2005, 17, 61- 76.
doi: 10.1105/tpc.104.026161 |
|
|
Ren P , Meng Y , Li B , et al. Molecular mechanisms of acclimatization to phosphorus starvation and recovery underlying full length transcriptome profiling in barley(Hordeum vulgare L. ). Frontiers in Plant Science, 2018, 9, 500.
doi: 10.3389/fpls.2018.00500 |
|
|
Rigamonte T A , Silveira W B , Fietto L G , et al. Restricted sugar uptake by sugar-induced internalization of the yeast lactose/galactose permease Lac12. FEMS Yeast Research, 2011, 11 (3): 243- 251.
doi: 10.1111/j.1567-1364.2010.00709.x |
|
|
Rottmann W H , Meilan R , Sheppard L A , et al. Diverse effects of overexpression of LEAFY and PTLF, a poplar(Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant Journal, 2000, 22 (3): 235- 245.
doi: 10.1046/j.1365-313x.2000.00734.x |
|
|
Ru°ži Dč ka K , Ursache R , Hejátko J , et al. Xylem development-from the cradle to the grave. New Phytologist, 2015, 207 (3): 519- 535.
doi: 10.1111/nph.13383 |
|
|
Sampedro J , Valdivia E R , Fraga P , et al. Soluble and membrane-bound β-glucosidases are involved in trimming the xyloglucan backbone. Plant Physiology, 2017, 173 (2): 1017- 1030.
doi: 10.1104/pp.16.01713 |
|
|
Samuga A , Joshi C P . Differential expression patterns of two new primary cell wall-related cellulose synthase cDNAs, PtrCesA6 and PtrCesA7 from aspen trees. Gene, 2004, 334, 73- 82.
doi: 10.1016/j.gene.2004.02.057 |
|
|
Sanchez C , Bauer S , Hematy K , et al. CHITINASE-LIKE1/POM-POM1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell, 2012, 24 (2): 589- 607.
doi: 10.1105/tpc.111.094672 |
|
|
Schoch G , Goepfert S , Morant M , et al. CYP98A3 from Arabidopsis thaliana is a 3'-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. Journal of Biological Chemistry, 2001, 276 (39): 36566- 36574.
doi: 10.1074/jbc.M104047200 |
|
|
Secco D , Jabnoune M , Walker H , et al. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell, 2013, 25 (11): 4285- 4304.
doi: 10.1105/tpc.113.117325 |
|
|
Sharon D , Tilgner H , Grubert F , et al. A single-molecule long-read survey of the human transcriptome. Nature Biotechnology, 2013, 31 (11): 1009- 1014.
doi: 10.1038/nbt.2705 |
|
|
Somerville R C , Chris R S . The cellulose synthase superfamily. Plant Physiology, 2000, 124 (2): 495- 498.
doi: 10.1104/pp.124.2.495 |
|
|
Song W , Jiang K , Zhang F , et al. Transcriptome sequencing, de novo assembly and differential gene expression analysis of the early development of Acipenser baeri. PLoS ONE, 2015, 10 (9): e0137450.
doi: 10.1371/journal.pone.0137450 |
|
|
Suzuki S , Li L , Sun Y H , et al. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiology, 2006, 142 (3): 1233- 1245.
doi: 10.1104/pp.106.086678 |
|
|
Tilgner H , Jahanbani F , Blauwkamp T , et al. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nature Biotechnology, 2015, 33 (7): 736- 742.
doi: 10.1038/nbt.3242 |
|
| Torre A R D L , Piot A , Liu B , et al. Functional and morphological evolution in gymnosperms: A portrait of implicated gene families. Evolutionary Applications, 2019, 13 (1): 210- 227. | |
|
Valério L , Meyer M D , Penel C , et al. Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry, 2004, 65 (10): 1331- 1342.
doi: 10.1016/j.phytochem.2004.04.017 |
|
|
Wang J P , Matthews M L , Naik P P , et al. Flux modeling for monolignol biosynthesis. Current Opinion in Biotechnology, 2019a, 56, 187- 192.
doi: 10.1016/j.copbio.2018.12.003 |
|
|
Wang L , Jiang X , Wang L , et al. A survey of transcriptome complexity using PacBio single-molecule real-time analysis combined with Illumina RNA sequencing for a better understanding of ricinoleic acid biosynthesis in Ricinus communis. BMC Genomics, 2019b, 20 (1): 456.
doi: 10.1186/s12864-019-5832-9 |
|
|
Xie L Q , Yang C J , Wang X L . Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. Journal of Experimental Botany, 2011, 62 (13): 4495- 4506.
doi: 10.1093/jxb/err164 |
|
|
Yan X , Liu J , Kim H , et al. CAD 1 and CCR 2 protein complex formation in monolignol biosynthesis in Populus trichocarpa. New Phytologist, 2019, 222 (1): 244- 260.
doi: 10.1111/nph.15505 |
|
|
Ye Z H , Zhong R . Molecular control of wood formation in trees. Journal of Experimental Botany, 2015, 66 (14): 4119- 4131.
doi: 10.1093/jxb/erv081 |
|
|
Young M D , Wakefield M J , Smyth G K , et al. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology, 2010, 11 (2): R14.
doi: 10.1186/gb-2010-11-2-r14 |
|
|
Zhang Y , Zhang S G , Han S Y , et al. Transcriptome profiling and in silico analysis of somatic embryos in Japanese larch(Larix leptolepis). Plant Cell Reports, 2012, 31 (9): 1637- 1657.
doi: 10.1007/s00299-012-1277-1 |
|
|
Zhong R Q , Lee C H , Zhou J L , et al. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell, 2008, 20 (10): 2763- 2782.
doi: 10.1105/tpc.108.061325 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||