

Scientia Silvae Sinicae ›› 2025, Vol. 61 ›› Issue (6): 109-119.doi: 10.11707/j.1001-7488.LYKX20240401
• Research papers • Previous Articles Next Articles
Yadi Wu1,2,Shu Diao1,Xianyin Ding1,Qinyun Huang1,Qifu Luan1,*( )
)
Received:2024-06-28
Online:2025-06-10
Published:2025-06-26
Contact:
Qifu Luan
E-mail:qifu.luan@caf.ac.cn
CLC Number:
Yadi Wu,Shu Diao,Xianyin Ding,Qinyun Huang,Qifu Luan. Construction of a DNA Fingerprinting of Elite Varieties of Introduced Exotic Pines in China Based on 51K Liquid-Phased Probes[J]. Scientia Silvae Sinicae, 2025, 61(6): 109-119.

Fig.1
Phylogenetic and population genetic analyses of 38 pine varieties A: The phylogenetic analysis. B: Genetic structure analysis of 38 pines at K=2. C: The Delta K line graph, with the peak at K=2 highlighting the optimal number of clusters for further genetic analysis. D: Principal component analysis, black dots represent slash pine, red dots represent loblolly pine, green dots represent P. elliottii × P. caribaea, and blue dots represent P. taeda × P. caribaea. PEE:湿地松Pinus elliottii;PTA:火炬松P. taeda;PAC:火加松P. taeda × P. caribaea;PEC:湿加松P. elliottii × P. caribaea. 英文缩写(PEE等)后面的数字是样本编号。The digits after the English abbreviation (PEE, etc.) are the sample number. K:最佳分群数Optimal number of groups."
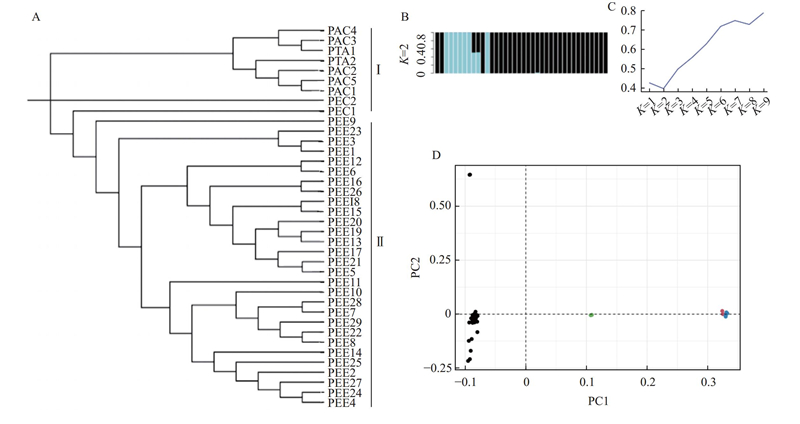
Table 2
Steps, criteria, and retained SNPs for core SNP selection"
| 筛选步骤 Screening steps | 筛选条件 Screening criteria | 剩余SNP的数量 Number of remaining SNPs |
| 原始数据 Raw data | — | 183 849 |
| 第1步 Step 1 | 次要等位基因频率大于0.3,缺失率为0 MAF >0.3,miss rate=0 | 22 593 |
| 第2步 Step 2 | 符合哈迪-温伯格平衡 HWE <1E-4 | 13 931 |
| 第3步 Step 3 | 连锁不平衡大于0.2 LD >0.2 | 8 502 |
| 第4步 Step 4 | 多态信息含量大于0.35 PIC >0.35 | 8 502 |
| 第5步 Step 5 | 杂合度小于0.25 He <0.25 | 344 |
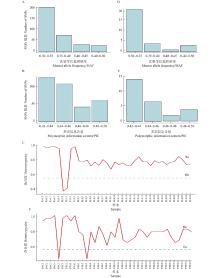
Fig.2
Genetic parameter information of 344 SNPs and 28 SNPs A–C: MAF (A), PIC (B),Ho and He (C) of 344 SNPs;D–F: MAF (D), PIC (E), Ho and He (F) of 20 SNPs. MAF: Minor allele frequency;PIC: Polymorphic information content;He: expected heterozygosity;Ho: Observed heterozygosity. PEE:湿地松 Pinus elliottii;PTA:火炬松P. taeda;PAC:火加松P. taeda × P. caribaea;PEC:湿加松P. elliottii × P. caribaea."
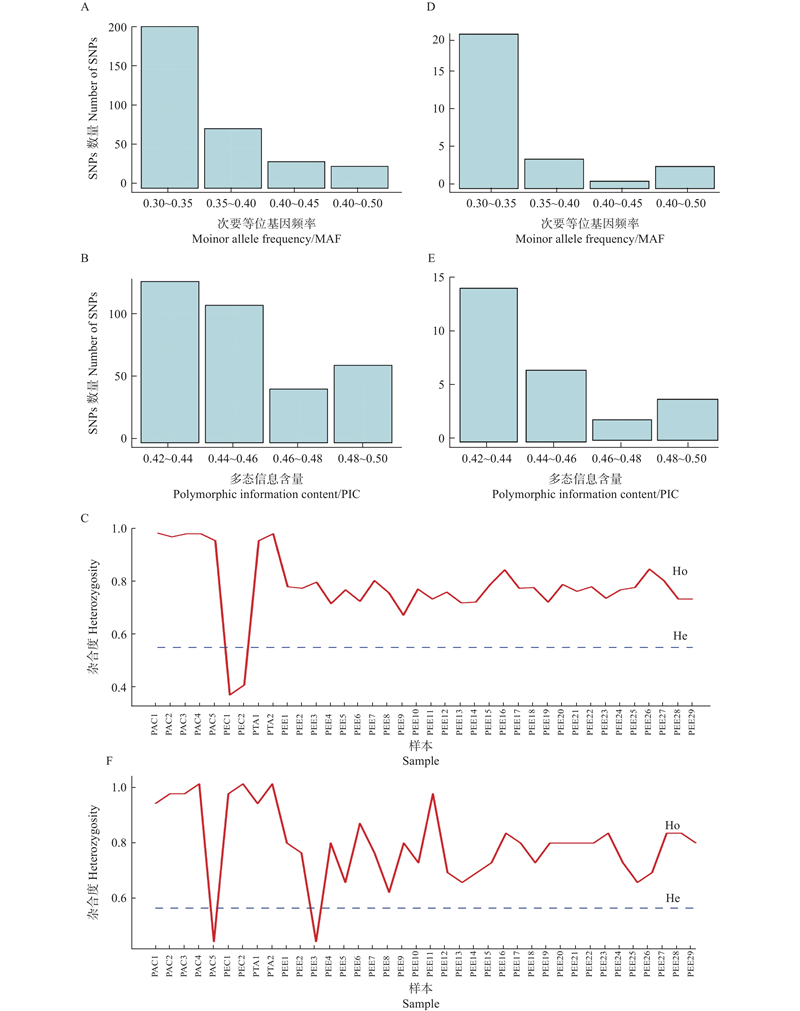
Table 4
20 SNP markers after simplification and genetic diversity index"
| 序号 No. | SNP名称 SNP ID | 染色体 Chromosome | 等位基因A Alley gene A | 等位基因B Alley gene B | 次要等位基 因频率 Minor allele frequency (MAF) | 多态信息含量 Polymorphic information content(PIC) |
| 1 | super2776.88036 | 10 | G | A | 0.500 0 | 0.500 0 |
| 2 | scaffold121890.3772 | 6 | T | C | 0.500 0 | 0.500 0 |
| 3 | scaffold124124.14341 | 11 | C | T | 0.500 0 | 0.500 0 |
| 4 | scaffold156626.9213 | 6 | A | G | 0.500 0 | 0.500 0 |
| 5 | Gene.280112.80 | 12 | C | T | 0.500 0 | 0.500 0 |
| 6 | super3477.582311 | 3 | G | A | 0.486 8 | 0.499 7 |
| 7 | super3658.67227 | 12 | G | C | 0.486 8 | 0.499 7 |
| 8 | super3673.196162 | 1 | G | C | 0.486 8 | 0.499 7 |
| 9 | scaffold4919.104166 | 11 | C | T | 0.486 8 | 0.499 7 |
| 10 | scaffold8912.56207 | 2 | C | T | 0.486 8 | 0.499 7 |
| 11 | scaffold18745.243173 | 7 | G | C | 0.486 8 | 0.499 7 |
| 12 | scaffold28210.166698 | 4 | C | T | 0.486 8 | 0.499 7 |
| 13 | scaffold45863.102243 | 8 | A | C | 0.486 8 | 0.499 7 |
| 14 | scaffold71374.123891 | 1 | G | T | 0.486 8 | 0.499 7 |
| 15 | scaffold120395.63994 | 12 | C | A | 0.486 8 | 0.499 7 |
| 16 | scaffold127392.228550 | 9 | A | C | 0.486 8 | 0.499 7 |
| 17 | scaffold144338.102562 | 10 | A | G | 0.486 8 | 0.499 7 |
| 18 | scaffold146854.112944 | 5 | A | T | 0.486 8 | 0.499 7 |
| 19 | Gene.117474.537 | 7 | T | C | 0.486 8 | 0.499 7 |
| 20 | super2715.43005 | 9 | C | T | 0.460 5 | 0.496 9 |

Fig.4
Phylogenetic relationship of 38 pines were analyzed based on 344 SNPs (A) and 20 SNPs (B) The black line represents Pinus elliottii (PEE) samples. The red line represents P. taeda (PTA) samples. The green line represents P. elliottii × P. caribaea (PEC) samples. The blue line represents P. taeda × P. caribaea (PAC) samples."
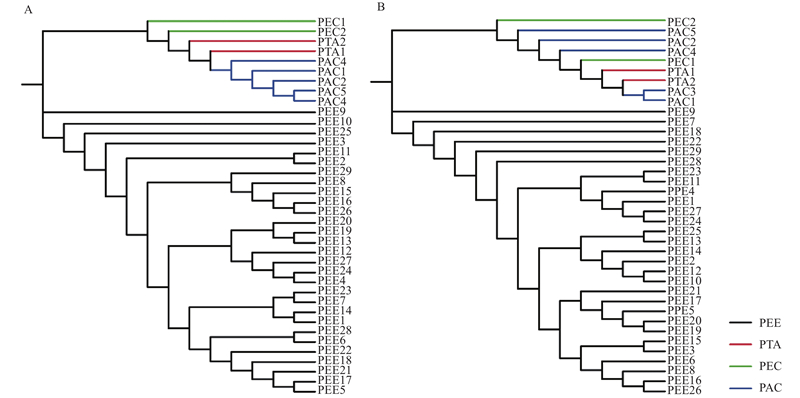
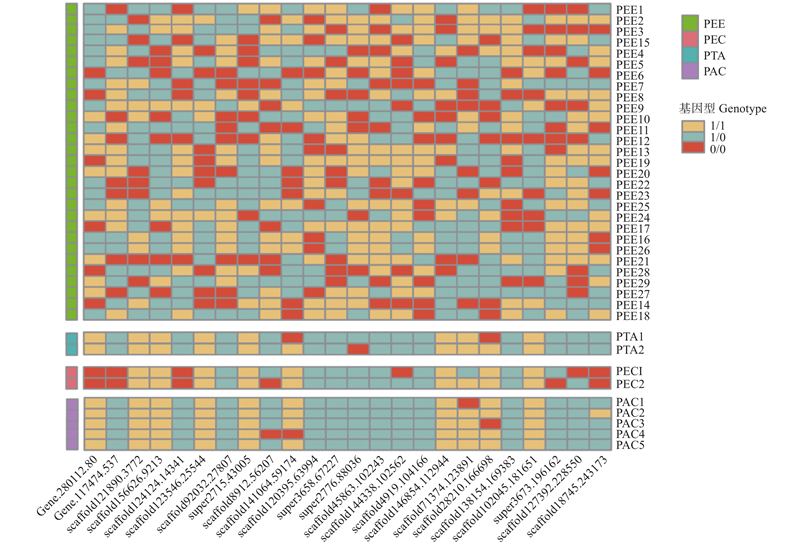
Fig.5
DNA fingerprinting of 38 pine germplasm resources Genotype is 1/1 represent that it is homozygote of major alle gene, and genotype is 0/0 represent that it is homozygote of minor alley gene, and genotype is 0/1 represent that it is heterozygote. The different color in the left represents different exotic pines species, the green plot line represents Pinus elliottii(PEE) samples. The blue plot represents P. taeda(PTA) samples. The pink plot represents P. elliottii × P. caribaea (PEC) samples. The purple plot represents P. taeda × P. caribaea (PAC) samples."
| 丁显印, 陶学雨, 刁 姝, 等. 2020. Pilodyn和Resistograph对湿地松活立木基本密度的评估. 南京林业大学学报(自然科学版), 44(3): 142−148. | |
| Ding X Y, Tao X Y, Diao S, et al. Estimation of wood basic density in a Pinus elliottii stand using Pilodyn and Resistograph measurements, Journal of Nanjing Forestry University (Natural Sciences Edition), 44(3): 142−148. [in Chinese] | |
|
樊晓静, 于文涛, 蔡春平, 等. 利用SNP标记构建茶树品种资源分子身份证. 中国农业科学, 2021, 54 (8): 1751- 1772.
doi: 10.3864/j.issn.0578-1752.2021.08.014 |
|
|
Fan X J, Yu W T, Cai C P, et al. Construction of molecular ID for tea cultivars by using of single-nucleotide polymorphism (SNP) markers. Scientia Agricultura Sinica, 2021, 54 (8): 1751- 1772.
doi: 10.3864/j.issn.0578-1752.2021.08.014 |
|
|
林 萍, 王开良, 姚小华, 等. 基于转录组SNP构建油茶主要品种资源的分子身份证. 中国农业科学, 2023, 56 (2): 217- 235.
doi: 10.3864/j.issn.0578-1752.2023.02.002 |
|
|
Lin P, Wang K L, Yao X H, et al. Development of DNA molecular ID in Camellia oleifera germplasm based on transcriptome-wide SNPs. Scientia Agricultura Sinica, 2023, 56 (2): 217- 235.
doi: 10.3864/j.issn.0578-1752.2023.02.002 |
|
| 刘超凡. 2022. 杨树无性系SSR指纹图谱及核心种质库构建. 长沙: 中南林业科技大学. | |
| Liu C F. 2022. Construction of SSR fingerprint database and core collection of Populus clones. Changsha: Central South University of Forestry & Technology. [in Chinese] | |
| 鲁仪增. 2019. 国槐遗传多样性评价及无性系分子鉴别. 北京: 中国林业科学研究院. | |
| Lu Y Z, 2019. Genetic diversity of Sophora japonica Linn. and clonal identification using molecular markers. Beijing: Chinese Academy of Forestry. [in Chinese] | |
|
栾启福, 姜景民, 张建忠, 等. 火炬松×加勒比松F_1代生长、树干通直度和基本密度遗传和配合力分析. 林业科学, 2011, 47 (3): 178- 183.
doi: 10.11707/j.1001-7488.20110327 |
|
|
Luan Q F, Jiang J M, Zhang J Z, et al. Estimation of heritability and combining ability for growth, stem-straightness and wood density of the F_1 generation of Pinus taeda × P. caribaea. Scientia Silvae Sinicae, 2011, 47 (3): 178- 183.
doi: 10.11707/j.1001-7488.20110327 |
|
| 栾启福, 李彦杰, 姜景民. 2014. 国外松种间杂交后代生长和形态性状变异及相关性分析. 植物研究, 34(1): 95–102. | |
| Luan Q F, Li Y J, Jiang J M, 2014. Variation of stem growth and morphology traits of exotic pine hybrids and their correlations. Bulletin of Botanical Research, 34(1): 95–102. [in Chinese] | |
| 栾启福. 2010. 几种松树杂交选育及其遗传分析. 北京: 中国林业科学研究院. | |
| Luan Q F. 2010. Hybridization of several pines and their genetical analysis. Beijing: Chinese Academy of Forestry. [in Chinese] | |
|
马庆国, 宋晓波, 贺君星, 等. 基于SSR分子标记的核桃种质资源分子身份证构建. 植物资源与环境学报, 2023, 32 (2): 1- 9.
doi: 10.3969/j.issn.1674-7895.2023.02.01 |
|
|
Ma Q G, Song X B, He J X, et al. Establishment of molecular identity cards of walnut (Juglans spp.) germplasm resources based on SSR molecular marker. Journal of Plant Resources and Environment, 2023, 32 (2): 1- 9.
doi: 10.3969/j.issn.1674-7895.2023.02.01 |
|
| 申 展. 2017. 基于形态学特性和SSR分子标记的文冠果种质资源评价与选优研究. 北京: 北京林业大学. | |
| Shen Z. 2017. Valuation and selection of Xanthoceras sorbifolium Bunge germplasm by morphology characteristics and SSR markers. Beijing: Beijing Forestry University. [in Chinese] | |
| 张良波. 2020. 光皮树优树群体遗传变异分析及核心种质构建. 北京: 北京林业大学. | |
| Zhang L B. 2020. Analysis of population genetic variation of plus trees and construction of core collection in Cornus wilsoniana. Beijing: Beijing Forestry University. [in Chinese] | |
| Batley J, Edwards D. 2007, SNP applications in plants//Oraguzie N C, Rikkerink E H A, Gardiner S E. eds. Association mapping in plants. New York, NY: Springer, 95−102. | |
|
Boban S, Maurya S, Jha Z. DNA fingerprinting: an overview on genetic diversity studies in the botanical taxa of Indian bamboo. Genetic Resource Crop Evolution, 2022, 69 (2): 469- 498.
doi: 10.1007/s10722-021-01280-8 |
|
|
Borders B E, Bailey R L. Loblolly pine—pushing the limits of growth. Southern Journal of Applied Forestry, 2001, 25 (2): 69- 74.
doi: 10.1093/sjaf/25.2.69 |
|
| Carvalho A, Matos M, Lima-Brito J, et al. DNA fingerprint of F1 interspecific hybrids from the Triticeae tribe using ISSRs. Euphytica, 2005, 143 (1/2): 93- 99. | |
|
Chambers G K, Curtis C, Millar C D, et al. DNA fingerprinting in zoology: past, present, future. Investigative Genetics, 2014, 5 (1): 3.
doi: 10.1186/2041-2223-5-3 |
|
|
Dallas J F. Detection of DNA “fingerprints” of cultivated rice by hybridization with a human minisatellite DNA probe. Proceedings of the National Academy of Sciences of the United States of America, 1988, 85 (18): 6831- 6835.
doi: 10.1073/pnas.85.18.6831 |
|
|
DeLaTorre, A R, Birol I, Bousquet J, et al. Insights into conifer Giga-genomes. Plant Physiology, 2014, 166 (4): 1724- 1732.
doi: 10.1104/pp.114.248708 |
|
|
DeVerno, L L, Mosseler A. Genetic variation in red pine (Pinus resinosa) revealed by RAPD and RAPD-RFLP analysis. Canadian Journal of Forest Research, 1997, 27 (8): 1316- 1320.
doi: 10.1139/x97-090 |
|
|
Diao S, Ding X Y, Luan Q F, et al. Development of 51 K liquid-phased probe array for Loblolly and Slash pines and its application to GWAS of Slash pine breeding population. Industrial Crops and Products, 2024, 216, 118777.
doi: 10.1016/j.indcrop.2024.118777 |
|
|
Ding X Y, Li Y J, Zhang Y N, et al. Genetic analysis and elite tree selection of the main resin components of Slash pine. Frontier Plant Science, 2023, 14, 1079952.
doi: 10.3389/fpls.2023.1079952 |
|
|
Duan Y F, Liu J, Xu J F, et al. DNA fingerprinting and genetic diversity analysis with simple sequence repeat markers of 217 potato cultivars (Solanum tuberosum L.) in China. American Journal of Potato Research, 2019, 96 (1): 21- 32.
doi: 10.1007/s12230-018-9685-6 |
|
| Ganal M W, Wieseke R, Luerssen H, et al. 2014. High-throughput SNP profiling of genetic resources in crop plants using genotyping arrays//Tuberosa R, Graner A, Frison E. eds. Genomics of plant genetic resources: volume 1. managing, sequencing and mining genetic resources. Dordrecht: Springer Netherlands, 113–130. | |
|
Ganea S, Ranade S S, Hall D, et al. Development and transferability of two multiplexes nSSR in Scots pine (Pinus sylvestris L.). Journal of Forestry Research, 2015, 26 (2): 361- 368.
doi: 10.1007/s11676-015-0042-z |
|
|
Geraldes A, DiFazio S P, Slavov G T, et al. A 34K SNP genotyping array for Populus trichocarpa: design, application to the study of natural populations and transferability to other Populus species. Molecular Ecology Resources, 2013, 13 (2): 306- 323.
doi: 10.1111/1755-0998.12056 |
|
|
Graham N, Telfer E, Frickey T, et al. Development and validation of a 36K SNP array for radiata pine (Pinus radiata D. Don). Forests, 2022, 13 (2): 176.
doi: 10.3390/f13020176 |
|
|
Guo Z F, Yang Q, Huang F F, et al. Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip. Plant Communications, 2021, 2 (6): 100230.
doi: 10.1016/j.xplc.2021.100230 |
|
|
Jackson C, Christie N, Reynolds S M, et al. A genome-wide SNP genotyping resource for tropical pine tree species. Molecular Ecology Resources, 2022, 22 (2): 695- 710.
doi: 10.1111/1755-0998.13484 |
|
| Jeffreys A J, Pena S D J. 1993. Brief introduction to human DNA fingerprinting//Pena S D J, Chakraborty R, Epplen J T, et al. eds. DNA fingerprinting: state of the science. Basel: Birkhäuser Verlag, 1–20. | |
|
Jeffreys A J, Wilson V, Thein S L. Hypervariable ‘minisatellite’ regions in human DNA. Nature, 1985, 314 (6006): 67- 73.
doi: 10.1038/314067a0 |
|
| Jehan T, Lakhanpaul S. Single nucleotide polymorphism (SNP)-methods and applications in plant genetics: a review. Indian Journal of Biotechnology, 2006, 5 (4): 435- 459. | |
| Jiang G L. 2015. Molecular marker-assisted breeding: a plant breeder’s review//Al-Khayri J M, Jain S M, Johnson D V. eds. Advances in plant breeding strategies: breeding, biotechnology and molecular tools. Cham: Springer International Publishing, 431–472. | |
| Kumar P, Gupta V K, Misra A K, et al. Potential of molecular markers in plant biotechnology. Plant Omics, 2020, 2 (4): 141- 162. | |
|
Lai M, Dong L M, Su R F, et al. Needle functional features in contrasting yield phenotypes of slash pine at three locations in southern China. Industrial Crops and Products, 2023, 206, 117613.
doi: 10.1016/j.indcrop.2023.117613 |
|
| Mammadov J, Aggarwal R, Buyyarapu R, et al. SNP markers and their impact on plant breeding. International Journal of Plant Genomics, 2012, 2012, 728398. | |
| Montanari S, Deng C, Koot E, et al. A multiplexed plant–animal SNP array for selective breeding and species conservation applications. G3 Genes| Genomes| Genetics, 2023, 13 (10): jkad170. | |
| Nadeem M A, Nawaz M A, Shahid M Q, et al. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnology & Biotechnological Equipment, 2018, 32 (2): 261- 285. | |
|
Nesbitt K A, Potts B M, Vaillancourt R E, et al. Partitioning and distribution of RAPD variation in a forest tree species, Eucalyptus globulus (Myrtaceae). Heredity, 1995, 74 (6): 628- 637.
doi: 10.1038/hdy.1995.86 |
|
|
Noormohammadi Z, Ibrahim-Khalili N, Sheidai M, et al. Genetic fingerprinting of diploid and tetraploid cotton cultivars by retrotransposon-based markers. Nucleus, 2018, 61 (2): 137- 143.
doi: 10.1007/s13237-018-0237-8 |
|
|
Nybom H, Weising K, Rotter B. DNA fingerprinting in botany: past, present, future. Investigative Genetics, 2014, 5 (1): 1.
doi: 10.1186/2041-2223-5-1 |
|
| Pavy N, Gagnon F, Rigault P, et al. Development of high-density SNP genotyping arrays for white spruce (Picea glauca) and transferability to subtropical and nordic congeners. Molecular Ecology Resources, 2013, 13 (2): 324- 336. | |
|
Prunier J, Verta J P, MacKay J J. Conifer genomics and adaptation: at the crossroads of genetic diversity and genome function. New Phytologist, 2016, 209 (1): 44- 62.
doi: 10.1111/nph.13565 |
|
| Rawat A, Barthwal S, Ginwal H S. Comparative assessment of SSR, ISSR and AFLP markers for characterization of selected genotypes of Himalayan Chir pine (Pinus roxburghii Sarg.) based on resin yield. Silvae Genetica, 2014, 63 (1): 94- 108. | |
|
Rodríguez S M, Ordás R, Alvarez J M. Conifer biotechnology: an overview. Forests, 2022, 13 (7): 1061.
doi: 10.3390/f13071061 |
|
|
Sousa-Santos C, Robalo J I, Collares-Pereira M J, et al. Heterozygous indels as useful tools in the reconstruction of DNA sequences and in the assessment of ploidy level and genomic constitution of hybrid organisms: Short Communication. DNA Sequence, 2005, 16 (6): 462- 467.
doi: 10.1080/10425170500356065 |
|
|
Tsumura Y, Yoshimura K, Tomaru N, et al. Molecular phytogeny of conifers using RFLP analysis of PCR-amplified specific chloroplast genes. Theoretical and Applied Genetics, 1995, 91 (8): 1222- 1236.
doi: 10.1007/BF00220933 |
|
|
Wang X R, Szmidt A E. Molecular markers in population genetics of forest trees. Scandinavian Journal of Forest Research, 2001, 16 (3): 199- 220.
doi: 10.1080/02827580118146 |
|
|
Wang Y Y, Lü H K, Xiang X H, et al. Construction of a SNP fingerprinting database and population genetic analysis of cigar tobacco germplasm resources in China. Frontiers in Plant Science, 2021, 12, 618133.
doi: 10.3389/fpls.2021.618133 |
|
|
Weiss-Schneeweiss H, Tremetsberger K, Schneeweiss G M, et al. Karyotype diversification and evolution in diploid and polyploid South American Hypochaeris (Asteraceae) inferred from rDNA localization and genetic fingerprint data. Annals of Botany, 2008, 101 (7): 909- 918.
doi: 10.1093/aob/mcn023 |
|
|
Wingett S W, Andrews S. FastQ screen: a tool for multi-genome mapping and quality control. F1000Research, 2018, 7, 1338.
doi: 10.12688/f1000research.15931.1 |
|
|
Wünsch A, Hormaza J I. Cultivar identification and genetic fingerprinting of temperate fruit tree species using DNA markers. Euphytica, 2002, 125 (1): 59- 67.
doi: 10.1023/A:1015723805293 |
|
|
Yang Y Y, Lyu M, Liu J, et al. Construction of an SNP fingerprinting database and population genetic analysis of 329 cauliflower cultivars. BMC Plant Biology, 2022, 22 (1): 522.
doi: 10.1186/s12870-022-03920-2 |
|
|
Zhang Y, Luan Q F, Jiang J J, et al. Prediction and utilization of malondialdehyde in exotic pine under drought stress using near-infrared spectroscopy. Frontiers in Plant Science, 2021, 12, 735275.
doi: 10.3389/fpls.2021.735275 |
|
|
Zhao D H, Bullock B P, Montes C R, et al. Loblolly pine outperforms slash pine in the southeastern United States – a long-term experimental comparison study. Forest Ecology and Management, 2019, 450, 117532.
doi: 10.1016/j.foreco.2019.117532 |
|
|
Zimin A, Stevens K A, Crepeau M W, et al. Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics, 2014, 196 (3): 875- 890.
doi: 10.1534/genetics.113.159715 |
| [1] | Li Wei, Yu Zhenzhen, He Hui, Zhao Jialu, Liu Xijun. Effects of Carbon Input Change on Soil Bacterial and Fungal Community Structure and Diversity in a Mature Pinus elliottii Plantation [J]. Scientia Silvae Sinicae, 2025, 61(6): 232-242. |
| [2] | Xiaoliang Che,Tianyi Liu,Zhe Wang,Ming Zeng,Quannian Li,Fencheng Zhao,Huishan Wu,Wenbing Guo. Effect of Different Stimulating Pastes on Oleoresin Yield of the Hybrid Pine (Pinus elliottii × P. caribaea) Plantation in Guangdong Province [J]. Scientia Silvae Sinicae, 2023, 59(8): 123-132. |
| [3] | Zhe Wang,Yang Liu,Fencheng Zhao,Ming Zeng,Fuming Li,Huishan Wu,Yiliang Li,Fangyan Liao,Leping Deng,Suiying Zhong,Wenbing Guo. Effects of Different Resin-Stimulant Pastes on Resin Production and Growth of Pinus elliottii Families [J]. Scientia Silvae Sinicae, 2022, 58(9): 106-116. |
| [4] | Huanying Fang,Shengsheng Xiao,Xiaofang Yu,Yong Xiong,Xunzhi Ouyang,Xiaolei Qin. Responses of Soil Respiration and Its Components to Simulated Acid Rain in Pinus elliottii Plantation [J]. Scientia Silvae Sinicae, 2021, 57(7): 20-31. |
| [5] | Linfeng Ye,Yan Li,Zhongyuan Wang,Shitong Lu,Tiantian Pan,Sen Chen,Jiangbo Xie. Efficiency-Safety Relationships of Hydraulic Conducting System for Branch and Root of Three Pinus Species Growing in Humid Area [J]. Scientia Silvae Sinicae, 2021, 57(7): 194-204. |
| [6] | Youming Xu,Caixia Zhou,Han Lin,Jiyun Tao,Juhua Zhang. Ultrastructural Changes of the Cambial Cells of Pinus elliottii during the Periods of Recovery Activity, Activity and Dormancy [J]. Scientia Silvae Sinicae, 2020, 56(10): 145-153. |
| [7] | Zhao Fencheng, Guo Wenbing, Zhong Suiying, Deng Leping, Wu Huishan, Lin Changming, Liao Fangyan, Tan Zhiqiang, Li Yiliang. Effects of Indirect Selection on Wood Density Based on Resistograph Measurement of Slash Pine [J]. Scientia Silvae Sinicae, 2018, 54(10): 172-179. |
| [8] | Zhang Shuainan, Luan Qifu, Jiang Jingmin. Genetic Variation Analysis for Growth and Wood Properties of Slash Pine Based on The Non-Destructive Testing Technologies [J]. Scientia Silvae Sinicae, 2017, 53(6): 30-36. |
| [9] | Luan Qifu;Lu Ping;Xiao Fuming;Jiang Jingmin;Yu Mukui. Investigation on the Damage of Pinus elliottii in the Freezing Rain and Snow Area and the Analysis on the Reason [J]. Scientia Silvae Sinicae, 2008, 44(11): 50-55. |
| [10] | Weng Yaofu;Chen Yuan;Zhao Yongchun;Zheng Kangle;Huang Shaofu;Zhang Jianzhong. DNA FINGERPRINTING OF CASTANEA MOLLISSIMA ELITE VARIETY (CLONE) SEEDLINGS [J]. Scientia Silvae Sinicae, 2001, 37(2): 51-55. |
| [11] | Jiangnan Li,Xirui Wan,Qiuping Zhong. MONITORING ON SOIL AND WATER EROSION IN DEMONSTRATION FOREST LAND OF SLASH PINE [J]. Scientia Silvae Sinicae, 1999, 35(zk): 71-73. |
| [12] | Mukui Yu,Hui Qiu,Liuyi Xu,Guiqin Wang,Xiaoyuan He. RESEARCH ON SOIL AND WATER LOSS LAW OF NEWLY-BUILT SAPLING FOREST LAND OF PINUS ELLIOTTII [J]. Scientia Silvae Sinicae, 1999, 35(zk): 20-28. |
| [13] | Huijun Jia,Huaiming Zheng,Jiangnan Li,Xirui Wan. STUDIES ON STEADY STATE MINERAL NUTRITION AND MINERALIZATION FOR CONTAINER-GROWN SEEDLINGS OF SLASH PINE [J]. Scientia Silvae Sinicae, 1998, 34(1): 9-17. |
| [14] | Huijun Jia,Huaiming Zheng,Xiaomei Hua,Jiangnan Li,Xirui Wan. EFFECTS OF STEADY-STATE MINERAL NUTRITION ON Pt ECTOMYCORRHIZAE FORMATION AND GROWTH OF CONTAINER-GROWN SEEDLINGS OF SLASH PINE [J]. Scientia Silvae Sinicae, 1997, 33(1): 51-58. |
| [15] | Xiaoming Wen,Xiuqin Luo,Ning Guan,Yunmin Song,Zhigang Pan. PRELIMINARY STUDIES ON RADIAL VARIATION PATTERNS OF WOOD PROPERTIES WITHIN TREES Ⅲ. STUDIES ON THE EFFECT OF TREE SPACING ON THE RADIAL VARIATION PATTERNS OF WOOD DENSITY IN SLASH PINE AND LOBLOLLY PINE [J]. Scientia Silvae Sinicae, 1996, 32(6): 536-542. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||