

Scientia Silvae Sinicae ›› 2020, Vol. 56 ›› Issue (10): 45-52.doi: 10.11707/j.1001-7488.20201005
Previous Articles Next Articles
Lei Zhang,Jianjun Hu*
Received:2019-10-30
Online:2020-10-25
Published:2020-11-26
Contact:
Jianjun Hu
CLC Number:
Lei Zhang,Jianjun Hu. An Analysis of T-DNA Insertion Loci and Detection of the Locus-Specific of Transgenic Populus nigra Lines with BtCry1Ac[J]. Scientia Silvae Sinicae, 2020, 56(10): 45-52.
Table 1
Primer sequences for insertion sites"
| 引物名称 Primer name | 引物序列 Primer sequence (5′—3′) |
| Bt-F | GAATTCGCTAGGAACCAAGCCATT |
| Bt-R | AAGTATATCCATCAAATGTGGACT |
| LAD1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA |
| LAD2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT |
| LAD3 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA |
| LAD4 | ACGATGGACTCCAGAGCGGCCGCBDNBNNNCGGT |
| AC1 | ACGATGGACTCCAGAG |
| LB-0A | CCCAACTTAATCGCCTTGCAGCACATC |
| LB-1A | ACGATGGACTCCAGTCCGGCCGATTTGGGTGATGG TTCACGTAGTGGG |
| LB-2A | TGGACCGCTTGCTGCAACTCTCTC |
| RB-0A | GGATACCGAGGGGAATTTATGGAACG |
| RB-1A | ACGATGGACTCCAGTCCGGCCGATAGTGACCTTAG GCGACTTTTGAACG |
| RB-2A | GTCATCGGCGGGGGTCATAACG |
| RB-F | TGTGGAGCATTGGTGTGAAG |
| RB-R | TTGAGTATGCCCTGCTGACA |
| LB-F | CCAACAATCCCCCAGATGAA |
| LB-R | AGGACACGGAAACCTTCGGA |
Table 2
Primer sequence for qRT-PCR"
| 引物名称 Primer name | 引物序列(正向) Primer sequence (Forward)(5′—3′) | 引物序列(反向) Primer sequence (Reverse)(5′—3′) |
| Actin | AAACTGTAATGGTCCTCCCTCCG | GCATCATCACAATCACTCTCCGA |
| qBt | TGTATGGGGACCGGATTCTA | CGAGCCTCGAAAACTACCAT |
| qSPK | TTTGGAACGATCACAGCTGC | GCTCTCCAGTTCCAGCACTA |
| qATM | AGGGGAAGGAATGCGAAAGA | GTCCCCAATGCCAACCTTG |
| qIBG | ACTCTACCAAGTCAGCCACC | AGGAGTGCATGGGAAAGTGA |
| qPIF1 | TGCCGGCTCTTAGAATTTGC | GACCCAAAGACTTGCAAGCA |
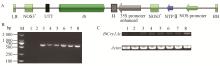
Fig.1
PCR detection of transgenic plants and expression level of BtCry1Ac A: T-DNA of vector pBIN437; B: PCR detection of transgenic poplar; C: Semi-quantitative PCR analysis of the BtCry1Ac. M: DL2000 DNA marker; 1-8: CK3, Populus deltoides 'Danhong', n12, n222, B3-44, B3-102, B3-132, B3-153 (B3:P. deltoides 'Danhong'×n222). "


Fig.2
Right boundary sequence of transgenic P. nigra n12 with BtCry1Ac A: The hiTAIL-PCR of right boundary in n12(M: DL2000 DNA marker; LAD1, LAD2, LAD3, LAD4 are degenerate primers; 2: The PCR products of AC1 and RB-1A; 3: The PCR products of AC1 and RB-2A); B: Right-boundary sequence and splicing position characteristics of n12. "
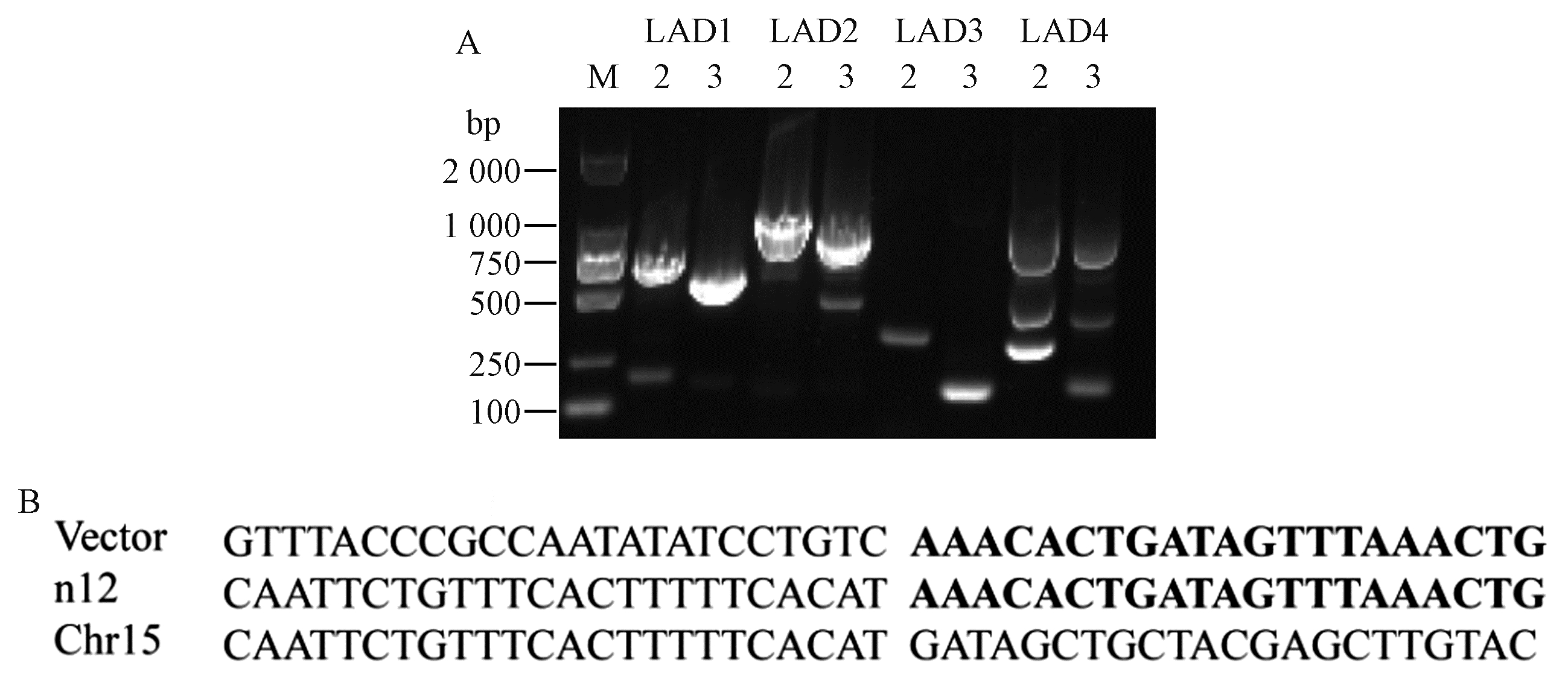

Fig.3
Left boundary sequence of transgenic P. nigra n222 A: The hiTAIL-PCR of left boundary in n222(M: DL2000 DNA marker; LAD1, LAD2, LAD3, LAD4 are degenerate primers; 2: The PCR products of AC1 and LB-1A; 3: The PCR products of AC1 and LB-2A); B: Left-boundary sequence and splicing position characteristics of n222. "
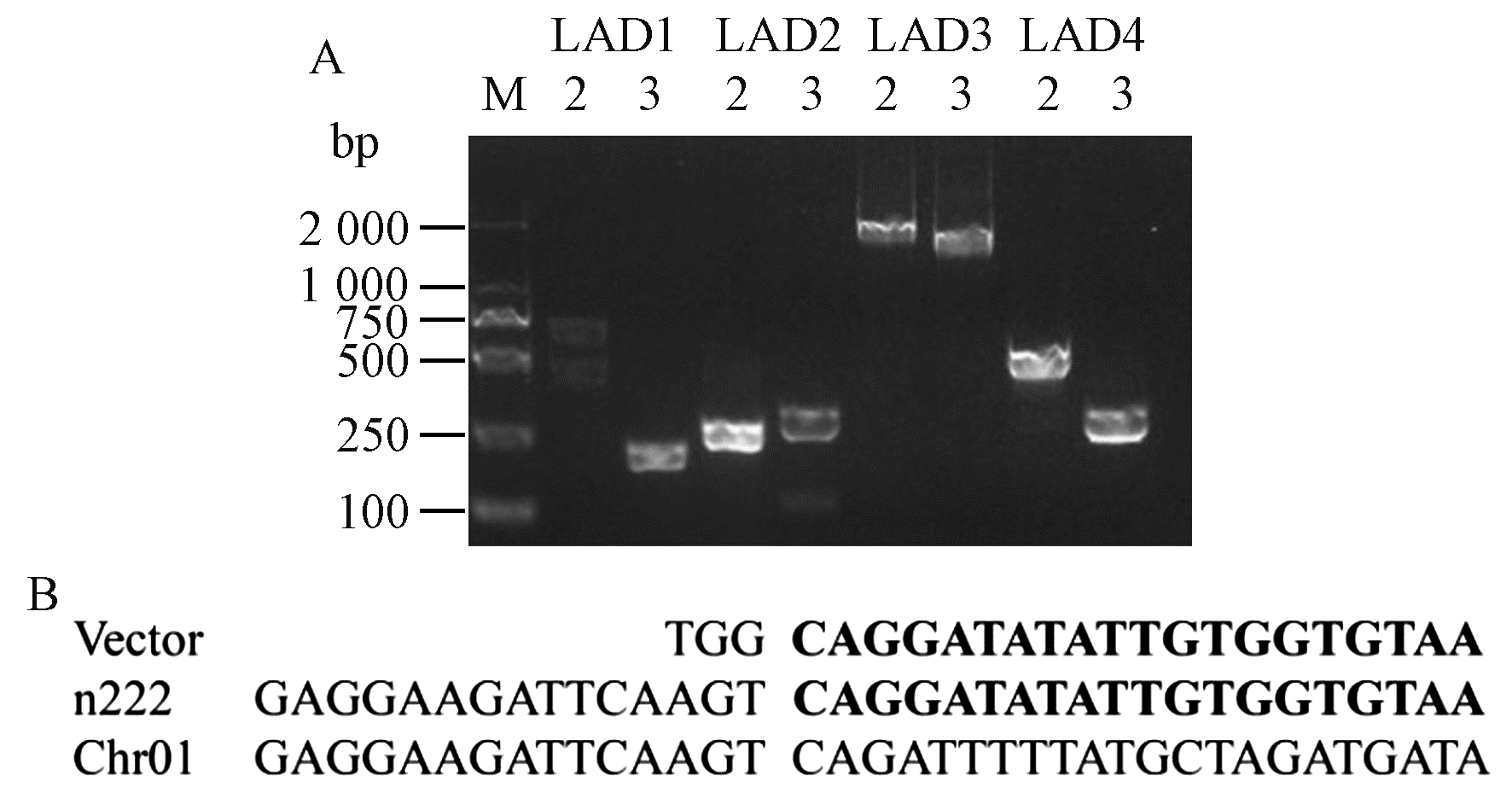
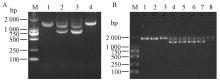
Fig.5
Specific PCR assay for insertion site of transgenic P. nigra n12 and n222 A: Specific PCR detection on right boundary in n12(M: DL2000 DNA marker; 1-4: CK3, n12, n12, n222); B: Specific PCR detection on left boundary in n222(M: DL2000 DNA marker; 1-8: CK3, P. deltoides 'Danhong', n12, n222, B3-44, B3-102, B3-132, B3-153). "
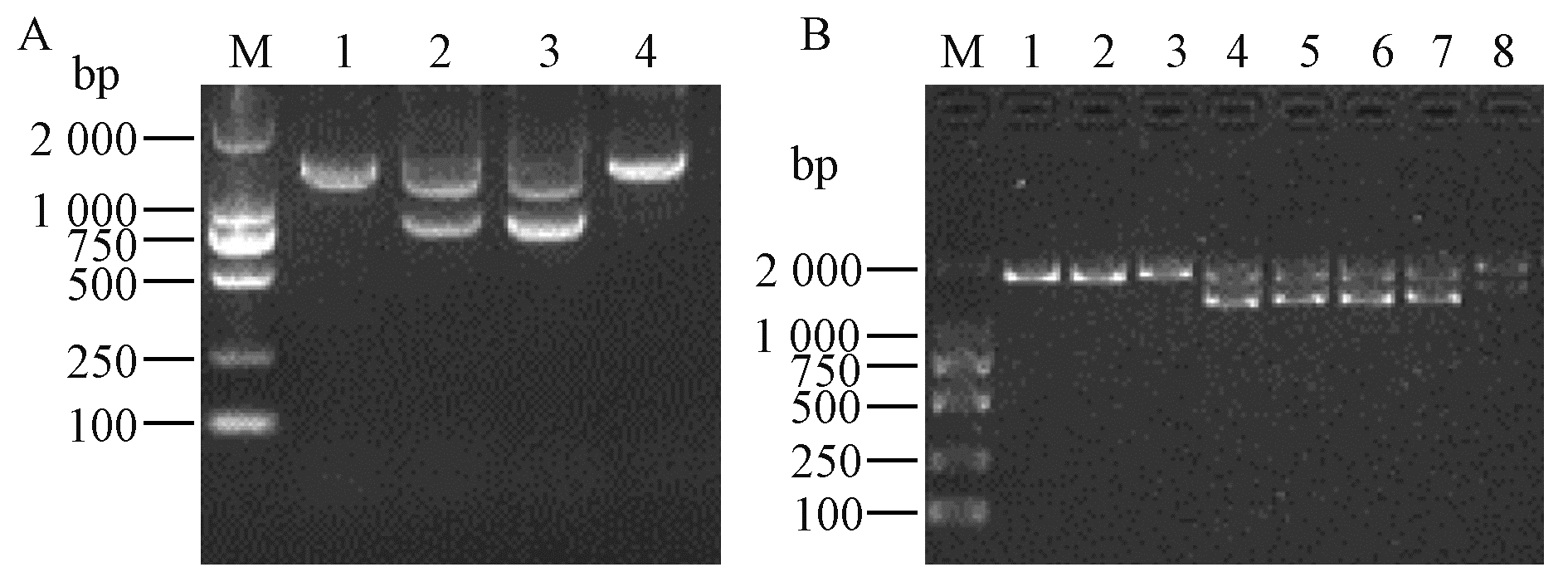
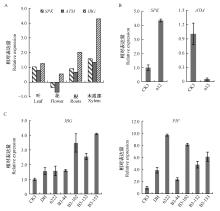
Fig.6
The expression levels of genes located at the T-DNA insertion sites A: Tissue specific expression of SPK, ATM and IBG; B: Relative expression level of SPK and ATM in transgenic P. nigra n12; C: Relative expression level of IBG and PIF in transgenic P. nigra n222 and hybrids. DH:P. deltoides 'Danhong'. "

| 蔡勤安, 尚丽霞, 姜志磊, 等. 转HAL1基因大豆侧翼序列分析及特异性检测方法研究. 大豆科学, 2018. 37 (1): 32- 38. | |
| Cai Q A , Shang L X , Jiang Z L , et al. The T-DNA flanking sequence analysis and specific detection method of transgenic soybean with gene HAL1. Soybean Science, 2018. 37 (1): 32- 38. | |
| 崔旭东. 2012.杨树抗旱多基因转化及整合机制研究.北京:中国林业科学研究院博士学位论文. | |
| Cui X D. 2012. Multi-drought resistance genes transformation and integration mechanism of poplar. Beijing: PhD thesis of Chinese Academy of Forestry.[in Chinese] | |
| 戴小枫, 郭予元. 我国棉花品种(系)抗虫性的鉴定、选育与应用. 植物保护学报, 1997. 24 (2): 179- 186. | |
| Dai X F , Guo Y Y . Current situation of the identification, selection, and application of resistant cotton cultivar against cotton insect pests in China. Journal of Plant Protection, 1997. 24 (2): 179- 186. | |
| 邓力华, 邓晓湘, 魏岁军, 等. 抗虫抗除草剂转基因水稻B1C893的获得与鉴定. 杂交水稻, 2014. 29 (1): 67- 75. | |
| Deng L H , Deng X X , Wei S J , et al. Development and identification of herbicide and insect resistant transgenic plant B1C893 in rice. Hybrid Rice, 2014. 29 (1): 67- 75. | |
| 冮慧欣. 2019.白桦黄叶突变体的鉴定及BpGLK1功能研究.哈尔滨:东北林业大学博士学位论文. | |
| Gang H X. 2019. Identification of a clorina mutant in Betula platyphylla×B. pendula and function research of BpGLK1. Harbin: PhD thesis of Northeast Forestry University.[in Chinese] | |
| 郭超, 何行健, 邓力华, 等. 转基因水稻BarKasalath-01事件特异性检测. 分子植物育种, 2017. 15 (11): 4466- 4475. | |
| Guo C , He X J , Deng L H , et al. Event-specific detection of genetically modified rice BarKasalath-01. Molecular Plant Breeding, 2017. 15 (11): 4466- 4475. | |
| 胡建军, 卢孟柱, 杨敏生. 我国抗虫转基因杨树生态安全性研究进展. 生物多样性, 2010. 18 (4): 336- 345. | |
| Hu J J , Lu M Z , Yang M S . Advances in biosafety studies on transgenic insect-resistant poplars in China. Biodiversity Science, 2010. 18 (4): 336- 345. | |
| 贾会霞, 孙佩, 李建波, 等. 丹红杨×转BtCry1Ac欧洲黑杨杂交子代抗虫性及生长量测定. 分子植物育种, 2017. 15 (10): 4101- 4109. | |
| Jia H X , Sun P , Li J B , et al. Insect-resistance and growth measurement of hybrid progeny from Populus deltoides 'Danhong' and transgenic P. nigra with BtCry1Ac gene. Molecular Plant Breeding, 2017. 15 (10): 4101- 4109. | |
| 刘航, 卢寰, 罗利, 等. 水稻剑叶突变体的表型分析及T-DNA旁邻基因分析. 植物科学学报, 2017. 35 (5): 708- 715. | |
| Liu H , Lu H , Luo L , et al. Phenotypic analysis of a rice flag leaf mutant and T-DNA flanking genes. Plant Science Journal, 2017. 35 (5): 708- 715. | |
| 申爱娟, 陈松, 周晓婴, 等. 转基因油菜W-4 T-DNA旁侧序列分析与事件特异性检测. 江苏农业学报, 2014. 30 (1): 14- 20. | |
| Shen A J , Chen S , Zhou X Y , et al. Analysis of the flanking sequence and event-specific detection of transgenic line W-4 of Brassica napus. Jiangsu Journal of Agricultural Sciences, 2014. 30 (1): 14- 20. | |
| 田颖川, 秦晓峰, 许丙寅, 等. 表达苏云金杆菌δ-内毒素基因的转基因烟草的抗虫性. 生物工程学报, 1991. 7 (1): 1- 10. | |
| Tian Y C , Qin X F , Xu B Y , et al. Insect resistance of transgenic tobacco plants expressing δ-endotoxin gene of Bacillus thuringiensis. Chinese Journal of Biotechnology, 1991. 7 (1): 1- 10. | |
| 田颖川. 抗虫转基因欧洲黑杨的培育. 生物工程学报, 1993. 9 (4): 291- 297. | |
| Tian Y C . Insect tolerance of transgenic Populus nigra plants transformed with Bacillus thuringiensis toxin gene. Chinese Journal of Biotechnology, 1993. 9 (4): 291- 297. | |
| 王晓波, 蒋凌雪, 魏利, 等. 外源抗草甘膦EPSPs基因在大豆基因组中的整合与定位. 作物学报, 2010. 36 (3): 365- 375. | |
| Wang X B , Jiang L X , Wei L , et al. Integration and insertion site of EPSPs gene on the soybean genome in genetically modified glyphosate-resistant soybean. Acta Agronomica Sinica, 2010. 36 (3): 365- 375. | |
| 魏岁军. 2014.转基因水稻EB7001S和BlC893的分子特征鉴定和相关性状评价.长沙:中国科学院大学硕士学位论文. | |
| Wei S J. 2014. Molecular identification and evaluation of transgenic rice EB7001S and BlC893. Changsha: MS thesis of University of Chinese Academy of Sciences.[in Chinese] | |
| 许文涛, 白卫滨, 罗云波, 等. 转基因产品检测技术研究进展. 农业生物技术学报, 2008. 16 (4): 714- 722. | |
| Xu W T , Bai W B , Luo Y B , et al. Progress in detection technique for genetically modified organisms. Journal of Agricultural Biotechnology, 2008. 16 (4): 714- 722. | |
| 杨琳, 付凤玲, 李晚忱. 农杆菌介导转基因植物T-DNA的整合方式. 遗传, 2011. 33 (12): 1327- 1334. | |
| Yang L , Fu F L , Li W C . T-DNA integration patterns in transgenic plants mediated by Agrobacterium tumefaciens. Hereditas, 2011. 33 (12): 1327- 1334. | |
| 张春玲, 李淑梅, 赵自成, 等. 杨树新品种'丹红杨'. 林业科学, 2008. 44 (1): 169. | |
| Zhang C L , Li S M , Zhao Z C , et al. A new poplar variety Populus deltoides Cl. 'Danhong'. Scientia Silvae Sinicae, 2008. 44 (1): 169. | |
| Barakat A , Gallois P , Raynal M , et al. The distribution of T-DNA in the genomes of transgenic Arabidopsis and rice. FEBS Letters, 2000. 471 (2/3): 161- 164. | |
|
Hilbeck A , Defarge N , Bøhn T , et al. Impact of antibiotics on efficacy of Cry toxins produced in two different genetically modified Bt maize varieties in two Lepidopteran herbivore species, Ostrinia nubilalis and Spodoptera littoralis. Toxins (Basel), 2018. 10 (12): 489.
doi: 10.3390/toxins10120489 |
|
|
Hirakawa T , Katagiri Y , Ando T , et al. DNA double-strand breaks alter the spatial arrangement of homologous loci in plant cell. Scientific Reports, 2015. 5 (1): 11058.
doi: 10.1038/srep11058 |
|
|
Hu J J , Tian Y C , Han Y F , et al. Field evaluation of insect-resistant transgenic Populus nigra trees. Euphytica, 2001. 121 (2): 123- 127.
doi: 10.1023/A:1012015709363 |
|
| Hu J J, Wang L, Yan D, et al. 2014. Research and application of transgenic poplar in China//Fenning T. Forestry Sciences Vol. 81: Challenges and Opportunities for the World's Forests in the 21st Century. Dordrecht: Springer, 567-584. https://www.springer.com/series/5991. | |
|
Hu J J , Zhang J , Chen X , et al. An empirical assessment of transgene flow from a Bt transgenic poplar plantation. PLoS ONE, 2017. 12 (1): e0170201.
doi: 10.1371/journal.pone.0170201 |
|
| Kumar S , Fladung M . Transgene repeats in aspen:molecular characterisation suggests simultaneous integration of independent T-DNAs into receptive hotspots in the host genome. Molecular and General Genetics, 2000. 264 (1): 20- 28. | |
|
Liu Y G , Chen Y . High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequence. BioTechniques, 2007. 43 (5): 649- 656.
doi: 10.2144/000112601 |
|
| Nacry P , Camilleri C , Courtial B , et al. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics, 1998. 149 (2): 641- 650. | |
|
Szabados L , Kovács I , Oberschall A , et al. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. The Plant Journal, 2002. 32 (2): 233- 242.
doi: 10.1046/j.1365-313X.2002.01417.x |
|
| Vachon V , Laprade R , Schwartz J L . Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins:A critical review. Journal of Invertebrate Pathology, 2012. 11 (1): 1- 12. | |
|
Wang G , Castiglione S , Chen Y , et al. Poplar (Populus nigra L.) plants transformed with a Bacillus thuringiensis toxin gene:insecticidal activity and genomic analysis. Transgenic Research, 1996. 5 (5): 289- 301.
doi: 10.1007/BF01968939 |
|
|
Whiteley H R , Schnepf H E . The molecular biology of parasporal crystal body formation in Bacillus thuringiensis. Annual Review of Microbiology, 1986. 40 (1): 549- 576.
doi: 10.1146/annurev.mi.40.100186.003001 |
|
|
Yong L , Mario G R , Bekir U , et al. Analysis of T-DNA insertion site distribution patterns in Arabidopsis thaliana reveals special features of genes without insertions. Genomics, 2006. 87 (5): 645- 652.
doi: 10.1016/j.ygeno.2005.12.010 |
|
|
Yoshiyama K , Sakaguchi K , Kimura S . DNA damage response in plants:conserved and variable response compared to animals. Biology, 2013. 2 (4): 1338- 1356.
doi: 10.3390/biology2041338 |
|
|
Zhang J , Zhan T Y , Jia H X , et al. Whole-genome re-sequencing reveals molecular mechanisms of biomass changes in 11-year-old Bt transgenic poplar. Trees, 2018. 32, 1609- 1620.
doi: 10.1007/s00468-018-1737-5 |
|
|
Zhang N , Xu W , Bai W , et al. Event-specific qualitative and quantitative PCR detection of LY038 maize in mixed samples. Food Control, 2011. 22 (8): 1287- 1295.
doi: 10.1016/j.foodcont.2011.01.030 |
|
|
Zhong Y Y , Ahmed S , Deng G F , et al. Improved insect resistance against Spodoptera litura in transgenic sweet potato by overexpressing Cry1Aa toxin. Plant Cell Reports, 2019. 38, 1439- 1448.
doi: 10.1007/s00299-019-02460-8 |
| [1] | Weibo Sun,Xindong Gong,Yan Zhou,Hongyan Li. Photosynthetic Characteristics of Transgenic Poplars with Maize PEPC and PPDK Gene at Young Plant Stage [J]. Scientia Silvae Sinicae, 2020, 56(7): 33-43. |
| [2] | Xing Wu,Xingfeng Hu,Peizhen Chen,Xiaobo Sun,Fan Wu,Kongshu Ji. Cloning and Functional Analysis of PmPIN1 Gene from Pinus massoniana [J]. Scientia Silvae Sinicae, 2020, 56(3): 184-192. |
| [3] | Sun Weibo, Wei Zhaoqiong, Ma Xiaoxing, Wei Hui, Zhuge Qiang. Safety Assessment of a Field Trial of Three Types of Transgenic Poplar Nanlin895 [J]. Scientia Silvae Sinicae, 2020, 56(10): 53-62. |
| [4] | Zhang Chao, Wang Jinmao, Zhao Jie, Pang Dingwei, Zhang Dejian, Yang Minsheng. Expression Characteristics of Bt Gene in Transgenic Poplar Transformed by Different Multi-Gene Vectors [J]. Scientia Silvae Sinicae, 2019, 55(9): 61-70. |
| [5] | Zhang Qin, Xu Zongda, Zhao Kai, Li Xiaowei, Zhang Luosha, Zhang Qixiang. Isolation and Biological Function Analysis of Anthocyanin Regulatory Gene PmMYB1 from Prunus mume [J]. Scientia Silvae Sinicae, 2018, 54(10): 64-72. |
| [6] | Jiang Wenhu, Zhang Dejian, Liu Junxia, Li Chaoli, Lu Zhanyuan, Yang Minsheng. Comparative Analysis of Arthropod Communities in Transgenic Bt and Non-Transgenic Poplar-Cotton Composite Systems [J]. Scientia Silvae Sinicae, 2018, 54(10): 73-79. |
| [7] | Chen Panfei, Zuo Lihui, Wang Guiying, Wang Jinmao, Ren Yachao, Yang Minsheng. Growth and Physiological Responses of Transgenic Populus×euramericana cv. ‘4/76’ with Multiple Genes Under Salt Stress [J]. Scientia Silvae Sinicae, 2017, 53(7): 45-53. |
| [8] | Huang Juan, Chen Cun, Zhang Weixi, Ding Changjun, Su Xiaohua, Huang Qinjun. Effects of Drought Stress on Anatomical Structure and Photosynthetic Characteristics of Transgenic JERF36 Populus alba×P. berolinensis Seedling Leaves [J]. Scientia Silvae Sinicae, 2017, 53(5): 8-15. |
| [9] | Yin Wu, Sun Weibo, Zhou Yan, Zhuge Qiang. Cloning and Functional Analysis of Rubisco Activase Gene from Populus trichocarpa [J]. Scientia Silvae Sinicae, 2017, 53(4): 83-95. |
| [10] | Chen Panfei, Ren Yachao, Zhang Jun, Wang Jinmao, Yang Minsheng. Expression and Transportation of Bt Toxic Protein in 8-Year-Old Grafted Transgenic Poplar [J]. Scientia Silvae Sinicae, 2016, 52(7): 46-52. |
| [11] | Zhang Yiwen, Ren Yachao, Liu Jiaojiao, Liang Haiyong, Yang Minsheng. Exogenous Gene Expression on Transgenic Populus×euramericana cv. '74/76' Carrying Bivalent Insect-Resistant Genes [J]. Scientia Silvae Sinicae, 2015, 51(12): 45-52. |
| [12] | Zhu Wenxu, Zhang Bingyu, Huang Qinjun, Chu Yanguang, Ding Changjun, Zhang Weixi, Su Xiaohua. Effects of Multi-Gene Transgenic Populus×euramericana 'Guariento' on the Function of Microbial Population in the Rhizosphere Soil [J]. Scientia Silvae Sinicae, 2015, 51(11): 69-75. |
| [13] | Sun Haiwei;Liu Jing;Song Jian;Weng Manli;Luo Lei;Huang Yanyan;Zhang Hong;Niu Qinglin;Wang Bin;Feng Dianqi;. Molecular Identification and Cold-Resistance Analysis of the AmGS Transgenic Photinia×fraseri ‘Red Robin’ Plants [J]. Scientia Silvae Sinicae, 2012, 48(7): 30-38. |
| [14] | Yin Wu;Li Lisha;Wang Like;Wang Mingxiu;Zhuge Qiang. Analysis of Photosynthetic Characteristics of Transgenic Poplars with Maize PEPC Gene [J]. Scientia Silvae Sinicae, 2012, 48(6): 63-71. |
| [15] | Niu Xiaoyun;Huang Dazhuang;Yang Minsheng;Li Xiaofen;Fu Xinshuang. Temporal and Spatial Expression of Bt Toxic Protein in Transgenic Btcry3A Hybrid Poplar 741 [J]. Scientia Silvae Sinicae, 2011, 47(12): 154-157. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||