

Scientia Silvae Sinicae ›› 2022, Vol. 58 ›› Issue (8): 136-148.doi: 10.11707/j.1001-7488.20220814
• Research papers • Previous Articles Next Articles
Zhi Li,Xiaoyan Xue,Zhen Liu,Qifei Cai,Xiaodong Geng,Jian Feng,Huina Zhou,Tao Zhang,Mingwan Li,Yanmei Wang*
Received:2021-12-09
Online:2022-08-25
Published:2022-12-19
Contact:
Yanmei Wang
CLC Number:
Zhi Li,Xiaoyan Xue,Zhen Liu,Qifei Cai,Xiaodong Geng,Jian Feng,Huina Zhou,Tao Zhang,Mingwan Li,Yanmei Wang. Analysis of Bacterial Community Structure, Diversity and Functional Prediction in Different Organs of Healthy and Diseased Idesia polycarpa Plants[J]. Scientia Silvae Sinicae, 2022, 58(8): 136-148.

Fig.2
Bacterial communities in various organ areas The bar chart on the leaf corresponds to the total number of OTUs in each group, the black circle on the right indicates unique and common condition among each group, and the column chart on the top indicates the corresponding cross size of the set."
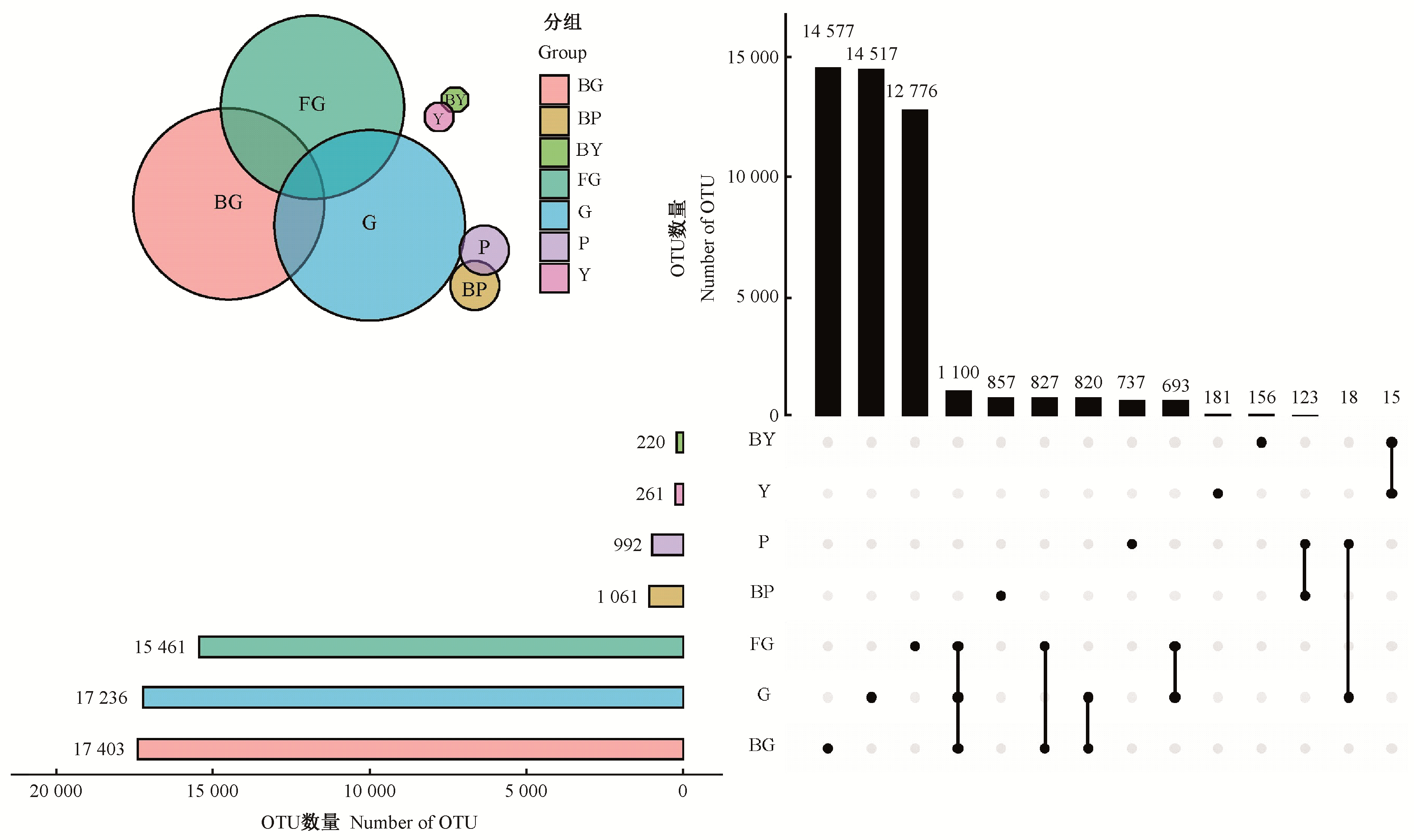
Table 1
Bacterial Alphadiversity index in different organ regions"
| 微生物 Microbial | 处理 Treatment | Chao1指数 Chao1 | 香农指数 Shannon | 辛普森指数 Simpson | 覆盖度 Goods coverage |
| 细菌 Bacteria | G | 6 435.47±664.00a | 11.79±0.14a | 0.999±0.000 07a | 0.983±0.009 7b |
| BG | 6 718.15±507.47a | 11.57±0.32a | 0.998±0.000 53a | 0.973±0.002 6c | |
| FG | 6 358.99±447.41a | 11.67±0.11a | 0.999±0.000 02a | 0.972±0.003 4c | |
| P | 427.49±116.28b | 3.06±0.43b | 0.612±0.042 84c | 0.999±0.000 5a | |
| BP | 414.14±125.19b | 3.50±0.86b | 0.752±0.084 34b | 0.998±0.000 6a | |
| Y | 117.78±34.15b | 0.79±0.06c | 0.268±0.053 04d | 0.999±0.000 3a | |
| BY | 90.19±3.42b | 0.63±0.16c | 0.208±0.089 40d | 0.999±0.000 1a |
Table 2
Abundance of main metabolic pathways in KEGG"
| 代谢通路 Metabolic pathway | 丰度 Abundance | |||||||
| 一级 Level 1 | 二级 Level 2 | G | BG | P | BP | Y | BY | |
| 细胞进程 Cellular processes | 细胞生长和死亡 Cell growth and death | 587.17 | 578.99 | 564.33 | 498.48 | 567.72 | 568.1 | |
| 细胞运动 Cell motility | 964.57 | 918.76 | 631.46 | 987.7 | 242.87 | 236.61 | ||
| 环境信息处理 Environmental information processing | 膜运输 Membrane transport | 702.54 | 717.12 | 656.2 | 840.38 | 606.78 | 604.78 | |
| 遗传信息处理 Genetic information processing | 折叠、分类和降解 Folding, sorting and degradation | 1 368.45 | 1 346.78 | 1 474.85 | 1 196.81 | 1 642.69 | 1 647.76 | |
| 复制和修复 Replication and repair | 2 132.13 | 2 084.86 | 1 804.58 | 1 722.23 | 1 796.45 | 1 797.94 | ||
| 翻译 Translation | 1 147.08 | 1 123.26 | 1 085.9 | 913.41 | 1 185.56 | 1 188.41 | ||
| 代谢 Metabolism | 氨基酸代谢 Amino acid metabolism | 5 794.02 | 5 726.99 | 4 552.88 | 4 328.21 | 4 303.87 | 4 296.93 | |
| 其他次生代谢产物的生物合成 Biosynthesis of other secondary metabolites | 1 271.03 | 1 268.49 | 821.18 | 741.07 | 791.38 | 804.33 | ||
| 碳水化合物代谢 Carbohydrate metabolism | 5 840.9 | 5 829.34 | 4 711.6 | 4 792.86 | 4 444.7 | 4 440.85 | ||
| 能量代谢 Energy metabolism | 2 221.1 | 2 199.72 | 3 034.14 | 2 390.84 | 3 500.67 | 3 513.25 | ||
| 聚糖生物合成与代谢 Glycan biosynthesis and metabolism | 1 158.34 | 1 188.84 | 1 062.5 | 952.41 | 1 082.64 | 1 082.92 | ||
| 脂质代谢 Lipid metabolism | 3 215.01 | 3 132.09 | 1 923.35 | 2 065.19 | 1 501.93 | 1 506.45 | ||
| 辅助因子和维生素的代谢 Metabolism of cofactors and vitamins | 5 058.61 | 4 993.13 | 5 618.58 | 4 713.01 | 6 123.86 | 6 123.65 | ||
| 其他氨基酸代谢 Metabolism of other amino acids | 3 477.05 | 3 464.45 | 3 380.19 | 3 187.17 | 3 329.89 | 3 333.09 | ||
| 萜类化合物和聚酮类化合物的代谢 Metabolism of terpenoids and polyketides | 4 257.25 | 4 239.98 | 2 950.06 | 2 943.4 | 2 651.67 | 2 593.58 | ||
| 核苷酸代谢 Nucleotide metabolism | 668.77 | 659.69 | 657.54 | 605.25 | 688.55 | 689.81 | ||
| 外源生物降解与代谢 Xenobiotics biodegradation and metabolism | 3 432.16 | 3 166.33 | 2 303.41 | 2 555.65 | 1 674.51 | 1 657.13 | ||
| 暴家兵, 齐果萍, 刘晋仙, 等. 华北落叶松树皮表面细菌群落多样性及其分布格局. 微生物学报, 2020, 60 (1): 135- 147. | |
| Bao J B , Qi G P , Liu J X , et al. Diversity and distribution pattern of bacterial community on bark surface of Larix principis-rupprechtii. Acta Microbiologica Sinica, 2020, 60 (1): 135- 147. | |
|
陈珺肄, 吴君, 房丽莎, 等. 温度与日长对山桐子萌芽成枝的影响. 西北林学院学报, 2019, 34 (3): 111- 117.111-117, 131
doi: 10.3969/j.issn.1001-7461.2019.03.17 |
|
|
Chen J Y , Wu J , Fang L S , et al. Effects of temperature and photoperiod on the branching after sprouted from buds in Idesia polycarpa. Journal of Northwest Forestry University, 2019, 34 (3): 111- 117.111-117, 131
doi: 10.3969/j.issn.1001-7461.2019.03.17 |
|
| 陈钊, 艾前进. 新中国明天的"空中油库"在这里诞生——油葡萄(山桐子)油品质、价值与市场前景分析论证会综述. 中国林业产业, 2019, (S2): 63- 67. | |
| Chen Z , Ai Q J . Tomorrow's "air oil depot" of new China is born here — a summary of the analysis and demonstration meeting on the quality, value and market prospect of oil grape (Idesia polycarpa) oil. China Forestry Industry, 2019, (2): 63- 67. | |
|
陈兆进, 林立安, 李英军, 等. 镉胁迫对芒草根际细菌群落结构、共发生网络和功能的影响. 环境科学, 2021, 42 (8): 3997- 4004.
doi: 10.13227/j.hjkx.202011198 |
|
|
Chen Z J , Lin L A , Li Y J , et al. Shifts in rhizosphere bacterial community structure, co-occurrence network, and function of Miscanthus after cadmium exposure. Environmental Science, 2021, 42 (8): 3997- 4004.
doi: 10.13227/j.hjkx.202011198 |
|
| 程浅. 烟草叶际细菌群落多样性与烟草野火病发生的关系及调控效应研究. 重庆: 西南大学, 2020. | |
| Cheng Q . Study on the relationship between the diversity of tobacco bacterial community and the occurrence of tobacco wildfire and its regulation effect. Chongqing: Southwest University, 2020. | |
| 丛微, 喻海茫, 于晶晶, 等. 人参种植对林地土壤细菌群落结构和代谢功能的影响. 生态学报, 2021, 41 (1): 162- 171. | |
| Cong W , Yu H M , Yu J J , et al. Effects of Ginseng cultivation on soil microbial community structure and metabolic functions in forest land. Acta Ecologica Sinica, 2021, 41 (1): 162- 171. | |
| 代莉. 山桐子种实地理变异研究. 郑州: 河南农业大学, 2014. | |
| Dai L . Study on geographic variation of fruit and seed of Idesia polycarpa Maxim. Zhengzhou: Henan Agricultural University, 2014. | |
| 丁钰珮, 杜宇佳, 高广磊, 等. 呼伦贝尔沙地樟子松人工林土壤细菌群落结构与功能预测. 生态学报, 2021, 41 (10): 1- 9. | |
| Ding Y P , Du Y J , Gao G L , et al. Soil bacterial community structure and functional prediction of Pinus sylvestris var. mongolica plantations in the HulunBuir Sandy Land. Acta Ecologica Sinica, 2021, 41 (10): 4131- 4139. | |
|
房丽莎, 徐自恒, 刘震, 等. 山桐子果实发育过程中内含物、内源激素及光合特性的变化. 林业科学, 2020, 56 (11): 41- 52.
doi: 10.11707/j.1001-7488.20201105 |
|
|
Fang L S , Xu Z H , Liu Z , et al. Changes of contents, endogenous hormones and photosynthetic characteristics of Idesia polycarpa fruit at the different developmental stages. Scientia Silvae Sinicae, 2020, 56 (11): 41- 52.
doi: 10.11707/j.1001-7488.20201105 |
|
| 葛艺, 徐民民, 徐绍辉, 等. 铜胁迫对小麦根系微域微生物群落的影响. 环境科学, 2021, 42 (2): 996- 1003. | |
| Ge Y , Xu M M , Xu S H , et al. Effects of copper pollution on microbial communities in wheat root system. Environmental Science, 2021, 42 (2): 996- 1003. | |
| 顾美英, 古丽尼沙·沙依木, 张志东, 等. 黑果枸杞不同组织内生细菌群落多样性. 微生物学报, 2021, 61 (1): 152- 166. | |
| Gu M Y , Gulinisha S Y M , Zhang Z D , et al. Diversity of endophytic bacterial communities in different tissues of Lycium ruthenicum. Acta Microbiologica Sinica, 2021, 61 (1): 152- 166. | |
| 李聪聪, 朱秉坚, 徐琳, 等. 高寒草甸优势植物叶内、根内与土壤原核微生物群落的分异. 生态学报, 2020, 40 (14): 4942- 4953. | |
| Li C C , Zhu B J , Xu L , et al. Differentiations of prokaryotic communities in leaf and root endosphere of dominant plants and bulk soils in alpinemeadows. Acta Ecologica Sinica, 2020, 40 (14): 4942- 4953. | |
| 李佛生, 司文瑾, 张踞林, 等. 山桐子内生真菌综合性实验的教学探索. 实验室科学, 2021, 24 (2): 49- 54. | |
| Li F S , Si W J , Zhang J L , et al. Teaching exploration on comprehensive experiments of Idesia polycarpa endophytic fungi. Laboratory Science, 2021, 24 (2): 49- 54. | |
| 李慧娟, 徐爱玲, 乔凤禄, 等. 冬青和女贞叶表面颗粒物微形态及叶际细菌群落结构. 环境科学, 2021, 42 (6): 3063- 3073. | |
| Li H J , Xu A L , Qiao F L , et al. Micro-morphological characteristics of particles on holly and ligustrum leaf surface and changes of bacterial community. Environmental Science, 2021, 42 (6): 3063- 3073. | |
| 林马水, 张媚, 柴莺飞, 等. 干腐病发病与健康山核桃树体pH值、养分与细菌多样性差异. 农业生物技术学报, 2019, 27 (2): 248- 259. | |
| Lin M S , Zhang M , Chai Y F , et al. Differences of pH values, nutrients and bacterial diversities between canker diseased and healthy Carya cathayensis trees. Journal of Agricultural Biotechnology, 2019, 27 (2): 248- 259. | |
| 刘海燕, 王敬敬, 赵维, 等. 塔里木河中下游流域棉田及胡杨林土壤细菌群落结构及多样性研究. 微生物学通报, 2019, 46 (9): 2214- 2230. | |
| Liu H Y , Wang J J , Zhao W , et al. Structure of soil bacteria community and diversity in cotton fieldand Euphrates poplar forest in the middle and lower reaches of Tarim river basin. Microbiology China, 2019, 46 (9): 2214- 2230. | |
| 马欣, 罗珠珠, 张耀全, 等. 黄土高原雨养区不同种植年限紫花苜蓿土壤细菌群落特征与生态功能预测. 草业学报, 2021, 30 (3): 54- 67. | |
| Ma X , Luo Z Z , Zhang Y Q , et al. Distribution characteristics and ecological function predictions of soil bacterial communities in rainfed alfalfa fields on the Loess Plateau. Acta Prataculturae Sinica, 2021, 30 (3): 54- 67. | |
| 南镇武, 刘柱, 代红翠, 等. 不同轮作休耕下潮土细菌群落结构特征. 环境科学, 2021, 42 (10): 4977- 4987. | |
| Nan Z W , Liu Z , Dai H C , et al. Characteristics of bacterial community structure in fluvo-aquic soil under different rotation fallow. Environmental Science, 2021, 42 (10): 4977- 4987. | |
| 秦立金, 徐峰, 刘永胜, 等. 黄瓜与西芹间作土壤细菌多样性及其对黄瓜枯萎病发生的影响. 中国生态农业学报, 2018, 26 (8): 1180- 1189. | |
| Qin L J , Xu F , Liu Y S , et al. Analysis of soil bacterial diversity under cucumber-celeryintercropping and its influence on cucumber Fusarium wilt. Chinese Journal of Eco-Agriculture, 2018, 26 (8): 1180- 1189. | |
| 涂昌, 柯文山, 邹路路, 等. 核桃细菌性疫病的生防菌筛选与鉴定. 北方园艺, 2020, (23): 10- 16. | |
| Tu C , Ke W S , Zou L L , et al. Screening and identification of biocontrol bacteria against bacterial blight of Juglans regia. Northern Horticulture, 2020, (23): 10- 16. | |
| 王安宁, 黄秋娴, 李晓刚, 等. 冀北山区不同植被恢复类型根际土壤细菌群落结构及多样性. 林业科学, 2019, 55 (9): 130- 141. | |
| Wang A N , Huang Q X , Li X G , et al. Bacterial community structure and diversity in rhizosphere soil of different vegetation restoration patterns in mountainous areas of northern Hebei. Scientia Silvae Sinicae, 2019, 55 (9): 130- 141. | |
| 王改萍, 阿依古丽·托乎提, 王茹, 等. 新疆乌尔禾地区盐渍土壤耐盐细菌多样性与群落结构研究. 微生物学杂志, 2021, 41 (2): 17- 26. | |
| Wang G P , Ayiguli T , Wang R , et al. Diversity and community structure of salt-tolerant bacteria in salinized soil in Wuerhe area in Xinjiang. Journal of Microbiology, 2021, 41 (2): 17- 26. | |
| 王文晓, 李小伟, 黄文广, 等. 蒙古沙冬青根际土壤细菌群落组成及多样性与生态因子相关性研究. 生态学报, 2020, 40 (23): 8660- 8671. | |
| Wang W X , Li X W , Huang W G , et al. Correlations between the composition and diversity of bacterial communities and ecological factors in the rhizosphere of Ammopiptanthus mongolicus. Acta Ecologica Sinica, 2020, 40 (23): 8660- 8671. | |
| 杨多, 岳海涛, 伍杰毅, 等. 胡杨叶片及韧皮部内生细菌多样性及生物学功能分析. 微生物学报, 2022, 62 (1): 213- 226. | |
| Yang D , Yue H T , Wu J Y , et al. Diversity and biological function of endophytic bacteria in Populus euphratica leaves and phloem. Acta Microbiologica Sinica, 2022, 62 (1): 213- 226. | |
| 杨莉. 欧美杨溃疡病病菌(Lonsdalea quercina)N-5-1致病相关基因的鉴定分析. 北京: 北京林业大学, 2013. | |
| Yang L . Identification and characterization of the pathogenicity-related genes in Lonsdalea quercina N-5-1. Beijing: Beijing Forestry University, 2013. | |
| 尹诗琳, 张建华, 李星月, 等. 桑轮纹病发生区桑叶表面细菌群落结构在冠层内的分异. 中国生态农业学报, 2021, 29 (3): 520- 530. | |
| Yin S L , Zhang J H , Li X Y , et al. Differentiation in the bacterial community structure of mulberry leaf surfaces in the canopy where mulberry ring rot disease occurs. Chinese Journal of Eco-Agriculture, 2021, 29 (3): 520- 530. | |
| 翟婉璐, 钟哲科, 高贵宾, 等. 覆盖经营对雷竹林土壤细菌群落结构演变及多样性的影响. 林业科学, 2017, 53 (9): 133- 142. | |
| Zhai W L , Zhong Z K , Gao G B , et al. Influence of mulching management on soil bacterial structure and diversity in Phyllostachys praecox stands. Scientia Silvae Sinicae, 2017, 53 (9): 133- 142. | |
| Amirsardari V , Sepahvand S , Madani M . Identification of deep bark canker agent of walnut and study of its phenotypic, pathogenic, holotypic and genetic diversity in Iran. Journal of Plant Interactions, 2017, 12 (1): 340- 347. | |
| Claesson M J , O'Sullivan O , Wang Q , et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE, 2009, 4 (8): e6669. | |
| Daniel B müller , Vogel C , Bai Y , et al. The plant microbiota: systems-level insights and perspectives. Annual Review of Genetics, 2016, 50 (1): 211- 234. | |
| Frutos D . Bacterial diseases of walnut and hazelnut and genetic resources. Journal of Plant Pathology, 2010, 92, S79- S85. | |
| Li Y , Zheng M , Wang H , et al. Brenneria corticis sp. nov., isolated from symptomatic bark of Populus×euramericana canker. International Journal of Systematic and Evolutionary Microbiology, 2019, 69 (1): 63- 67. | |
| Li Z , Siemann E , Deng B , et al. Soil microbial community responses to soil chemistry modifications inalpine meadows following human trampling. Catena, 2020, 194, 104717. | |
| Martine M , Hanneke H , Eric m . Brenneria salicis, the bacterium causing watermark disease in willow, resides as an endophyte in wood. Environmental Microbiology, 2009, 11 (6): 1453- 1462. | |
| Moretti C , Silvestri F M , Rossini E , et al. A protocol for rapid identification of Brenneria nigrifluens among bacteria isolated from bark cankers in Persian walnut plants. Journal of Plant Pathology, 2007, 89 (2): 211- 218. | |
| Müller D B , Vogel C , Bai Y , et al. The plant microbiota: systems-level insights and perspectives. Annual Review of Genetics, 2016, 50 (1): 211- 234. | |
| Qing T , Yong C L , Du X P , et al. Comparison of gut microbiota diversity and predicted functions between healthy and diseased captive Rana dybowskii. Frontiers in Microbiology, 2020, 11, 2096. | |
| Sun L , Hao Y , Wu Y M , et al. First report of alternarialeaf spot caused by Alternaria alternata on Idesia polycarpa in China. Plant Disease, 2015, 9 (8): 12- 13. | |
| Wu Y X , Qu M Q , Pu X H , et al. Distinct microbial communities among different tissues of citrus tree Citrus reticulata cv. Chachiensis. Scientific Reports, 2020, 10 (1): 6068. | |
| Zarraonaindia I , Owens S M , Weisenhorn P , et al. The soil microbiome influences grapevine-associated microbiota. MBio, 2015, 6 (2): e02527- 14. |
| [1] | Xiaolei Ding,Qingtong Wang,Sixi Lin,Ruiwen Zhao,Yue Zhang,Jianren Ye. Analysis of Genetic Variations of Bursaphelenchus xylophilus Populations between Guangdong and Jiangsu Provinces with SNP Marker [J]. Scientia Silvae Sinicae, 2022, 58(8): 1-9. |
| [2] | Shuzi Zhang,Jianting Yin,Qiwen Ren,Shubin Zhang,Xin Wang,Liandi Li,Jun Bi. Effect of Dominant Species on Diversity Pattern of Neighbor Species in Coniferous-Broadleaved Mixed Forest in Northern Hebei Mountains [J]. Scientia Silvae Sinicae, 2022, 58(4): 32-39. |
| [3] | Wenting Pan,Jianjun Sun,Qinqin Yuan,Lili Zhang,Kangqiao Deng,Yueqiao Li. Analysis of Genetic Diversity and Structure in Different Provenances of Liriodendron by RAD-seq Technique [J]. Scientia Silvae Sinicae, 2022, 58(4): 74-81. |
| [4] | Minxia Ren,Tan Li,Ziheng Zhang,Yuexia Zeng,Lifeng Wang,Minsheng Yang,Junxia Liu. Effects of Transgenic BtCry1Ac and API gene in Poplar 107 on Diversity and Stability of Arthropod Community [J]. Scientia Silvae Sinicae, 2022, 58(4): 110-118. |
| [5] | Shaoxian Huang,Hongxiang Wang,Hui Peng,Yaoyi Wang,Yuanfa Li,Shaoming Ye. Analysis of Second-Order Characteristics of Tree Species Dominance in an Old Growth Forest Community in Yachang Nature Reserve [J]. Scientia Silvae Sinicae, 2022, 58(4): 128-140. |
| [6] | Chen Liu,Chunyu Zhang,Xiuhai Zhao. Effects of Disturtance by Thinning on Productivity Stability of Conifer-Broadleaf Mixed Forest in Jiaohe, Jilin Province [J]. Scientia Silvae Sinicae, 2022, 58(3): 1-9. |
| [7] | Cong Li,Jinghua Lu,Mei Lu,Zhidong Yang,Pan Liu,Yulian Ren,Fan Du. Distribution of Soil Microbial Biomass Carbon and Nitrogen across Different Altitudinal Vegetation Zones in Wenshan National Nature Reserve [J]. Scientia Silvae Sinicae, 2022, 58(3): 20-30. |
| [8] | Feiran Jia,Zhongfu Zhou,Wenxia Zhao,Huiquan Sun,Yanxia Yao. Diversity of Gut Microorganisms in Natural Population of Agrilus mali (Coleoptera: Buprestidae) [J]. Scientia Silvae Sinicae, 2022, 58(3): 86-96. |
| [9] | Jingen Peng,Jinyu Gong,Yuhai Fan,Hua Zhang,Yinfeng Zhang,Yuqing Bai,Yanmei Wang,Lijuan Xie. Diversity of Soil Microbial Communities in Rhizosphere and Non-rhizosphere of Rhododendron moulmainense [J]. Scientia Silvae Sinicae, 2022, 58(2): 89-99. |
| [10] | Xuan Fang,Jingwei Wen,Yue Chen,Min Fan,Xingxia Ma. Fungal Diversity of Wooden Flume Unearthed from Nanyue National Palace Site under in situ Preservation Environment [J]. Scientia Silvae Sinicae, 2021, 57(7): 131-141. |
| [11] | Yanlin Zhang,Caifeng Huang,Mingzhuo Bao,Chuifan Zhou,Zongming He. Effects of Biochar and Its Aging Biochar on Soils Nutrients and Microbial Community Composition in Cunninghamia lanceolata Plantations: a Laboratory Simulation Experiment [J]. Scientia Silvae Sinicae, 2021, 57(6): 169-179. |
| [12] | Chaoqun Du,Xiaomei Sun,Yunhui Xie,Yimei Hou. Genetic Diversity of Larix kaempferi Populations with Different Levels of Improvement in Northern Subtropical Region [J]. Scientia Silvae Sinicae, 2021, 57(5): 68-76. |
| [13] | Xue Dong, Yonghua Li, Zhiming Xin, Ruibing Duan, bin Yao, Yanfeng Bao, Zhengguo Zhang, Yuan Liu. Patterns of Altitudinal Distribution of Species Diversity of Desert Gobi Shrub Communities in West Hexi Corridor of China [J]. Scientia Silvae Sinicae, 2021, 57(2): 168-178. |
| [14] | Jinyu Gong,Jingen Peng,Lijuan Xie,Yinfeng Zhang,Chaochan Li,Yanmei Wang. Microbial Diversity in Rhizosphere Soil of Rhododendron moulmainense with Different Tree Potential in Wutong Mountain of Shenzhen [J]. Scientia Silvae Sinicae, 2021, 57(11): 190-200. |
| [15] | Rongrong Pang,Jieying Peng,Yan Yan. Factors Influencing Aboveground Biomass in the Secondary Forest of Quercus aliena var. acutiserrata in Taibai Mountain [J]. Scientia Silvae Sinicae, 2021, 57(10): 157-165. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||