

Scientia Silvae Sinicae ›› 2023, Vol. 59 ›› Issue (5): 88-99.doi: 10.11707/j.1001-7488.LYKX20220397
Previous Articles Next Articles
Ke Yuan,Jianqin Huang,Ketao Wang,Guohua Xia,Qixiang Zhang,Chuanmei Xu*
Received:2022-06-12
Online:2023-05-25
Published:2023-08-02
Contact:
Chuanmei Xu
CLC Number:
Ke Yuan,Jianqin Huang,Ketao Wang,Guohua Xia,Qixiang Zhang,Chuanmei Xu. Development and Application of Oligonucleotide Probes for the Genus Carya[J]. Scientia Silvae Sinicae, 2023, 59(5): 88-99.
Table 1
Materials used for this study"
| 材料名称 Species name | 来源 Origin |
| 山核桃 C. cathayensis | 浙江省杭州市临安区 Lin'an District, Hangzhou City, Zhejiang Province |
| 大别山山核桃 C. dabieshanensis | 安徽省金寨县 Jinzhai County, Anhui Province |
| 云南山核桃 C. tonkinensis Lecte | 云南省红河州个旧市贾沙乡 Jiasha Township, Gejiu City, Honghe Prefecture, Yunnan Province |
| 湖南山核桃 C. hunanensis | 湖南省溆浦县龙潭镇 Longtan Town, Xupu County, Hunan Province |
| 贵州山核桃 C. kweichowensis | 贵州省安龙县德卧镇 Dewu Town, Anlong County, Guizhou Province |
| 喙核桃 C. sinensis | 贵州省望谟县大观镇 Daguan Town, Wangmo County, Guizhou Province |
| 薄壳山核桃 C. illinoinensis | 浙江农林大学校园 Zhejiang Agricultural and Forestry University Campus |
Table 2
The sequence and fluorescence modification information of six oligonucleotide probes"
| 探针名称Probes name | 序列信息及荧光修饰类型 The information of sequence and fluorescence modification |
| sht-2 | FAM-5'-GCCCGTGGGGCTTGTCAGGGTCAGGGGCAGTGTCAGGGCATG-3' |
| sht-3 | FAM-5'-CATGTGCCCGTGGGGCTTGTCAGGGTCAGGGGCAGTGTCAAGGGGCATGC-3' |
| sht-4 | FAM-5'-CAGTGTCAGGGCATGGCCCGTGGGGCTTGTCAGGGTCAGGGG-3' |
| sht-5 | FAM-5'-CAGGGGCAGTGTCAAGGGGCATGCCATGGTGCCCGTGGGGCTTGTCAAGGT-3' |
| sht-45S | TAMRA-5'-CGGGATTCTGCAATTCACACCAAGTATCGCATTTCGCTA-3' |
| TAMRA-5'-GAAACCTTGTTACGACTTCTCCTTCCTCTAAATGATA-3' | |
| TAMRA-5'-GCGACGGGCGGTGTGTACAAAGGGCAGGGACGTAGTCAA-3' | |
| TAMRA-5'-TTAACCAGACAAATCGCTCCACCAACTAAGAACGGCCATG-3' | |
| TAMRA-5'-TGCCCTTCCGTCAATTCCTTTAAGTTTCAGCCTTGCGACC-3' | |
| TAMRA-5'-TGGTCGGCATCGTTTATGGTTGAGACTAGGACGGTATCTG-3' | |
| TAMRA-5'-GGATTGGGTAATTTGCGCGCCTGCTGCCTTCCTTG-3' | |
| TAMRA-5'-TTCATACTTACACATGCATGGCTTAATCTTTGAGAC-3' | |
| sht-5s | FAM-5'-TGCGATCATACCAGCACCAATGCACCGGATCCCATTAGAACTCCGCAGTTAAGCGTG-3' |
| FAM-5'-TGCTTGGGCGAGAGTAGTACTAGGATGGGTGACCTCCTGGGAAGTCCTCGTGTTGCA-3' | |
| FAM-5'-TGCGATCGTACCAGCACTAATGCATCGGATCCCATCAGAACTCCGCAGTTAAGCGTG-3' |
Fig.5
Colocalization of oligo probe sht-2, sht-3, sht-4 and sht-5 on the chromsome of C. cathayensis a1 and a2 are sht-2 and sht-4, respectively. a3 is the colocalization of sht-2 and sht-4. b1 and b2 are sht-3 and sht-4, respectively. b3 is the colocalization of sht-3 and sht-4. c1 and c2 are sht-3 and sht-5, respectively. c3 is the colocalization of sht-3 and sht-5."
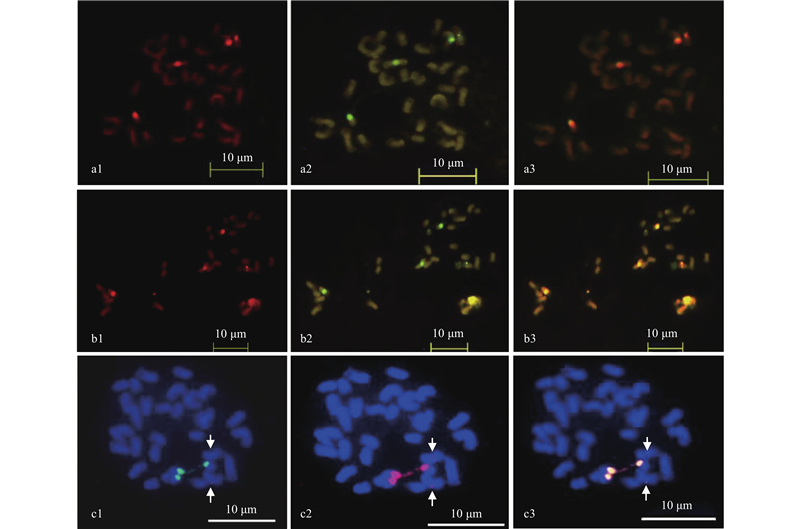
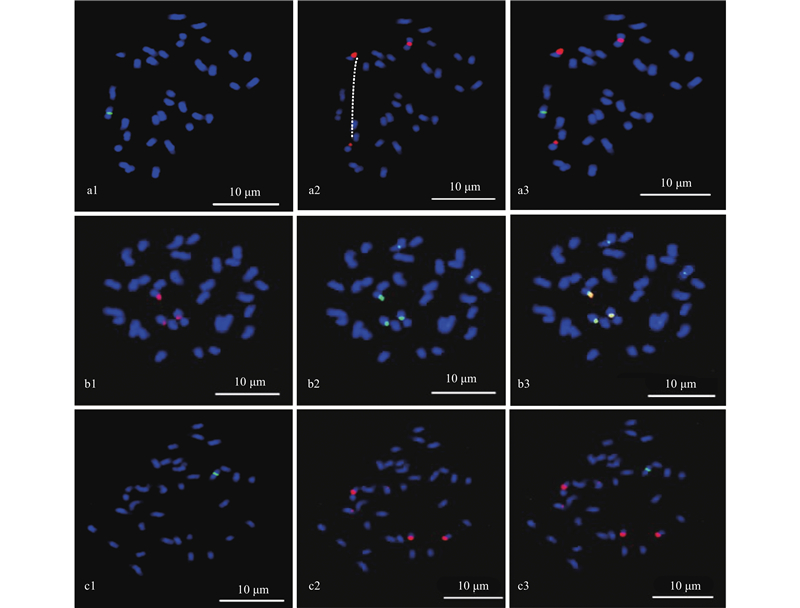
Fig.6
Colocalization of oligo probe sht-45S, sht-5S and sht-5 on the on the chromsome of C. cathayensis a1 and a2 are sht-5S (green) and sht-45S (red), respectively. a3 is the colocalization of sht-5S and sht-45S. b1 and b2 are sht-45S (red) and sht-5 (green), respectively. b3 is the colocalization of sht-45S and sht-5. c1 and c2 are sht-5S (green) and sht-5 (red), respectively. c3 is the colocalization of sht-5S and sht-5."
| 陈瑞阳, 李秀兰, 宋文芹, 等. 2003. 中国主要经济植物基因组染色体图谱Ⅳ. 北京: 科学出版社, 1−628. | |
| Chen R Y, Li X L, Song W Q, et al. 2003. Chromosome atlas of major economic plants genome in China (Version IV): Chromosome atlas of various bamboo species. Beijing: Science Press, 1−628.[in Chinese] | |
| 杜 培. 2017. 小麦、百萨偃麦草和花生染色体荧光原位杂交寡核苷酸探针(套)开发与应用. 南京: 南京农业大学. | |
| Du P. 2017. Development and application of wheat, thinopyrwn bessarabicwn and peanut oligonucleotide and multiplex probes for fluorescence in situ hybridization. Nanjing: Nanjing Agricultural University.[in Chinese] | |
| 刘茂春, 黎章矩. 中国山核桃属一新种. 浙江林学院学报, 1984, 1 (1): 41- 42. | |
| Liu M C, Li Z J. A new species of Carya from China . Journal of Zhejiang Forestry College, 1984, 1 (1): 41- 42. | |
| 吕芳德, 杨 帆, 张日清. 山核桃属部分种的核型分析. 中南林学院学报, 2002, 22 (1): 47- 49. | |
| Lü F D, Yang F, Zhang R Q. Karyotypes of three Carya Nutt. species . Journal of Central South Forestry University, 2002, 22 (1): 47- 49. | |
| 徐川梅. 2017. 10个山核桃种染色体核型及基因组类型分析. 北京: 中国林业科学研究院. | |
| Xu C M. 2017. Analysis of karyotype and genome type of ten Carya species. Beijing: Chinese Academy of Forestry.[in Chinese] | |
|
Albert P S, Zhang T, Semrau K, et al. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116 (5): 1679- 1685.
doi: 10.1073/pnas.1813957116 |
|
|
Braz G T, He L, Zhao H N, et al. Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics, 2018, 208 (2): 513- 523.
doi: 10.1534/genetics.117.300344 |
|
| Cai Z X, Liu H J, He Q Y, et al. Differential genome evolution and speciation of Coix lacryma-jobi L . and Coix aquatica Roxb. hybrid guangxi revealed by repetitive sequence analysis and fine karyotyping. BMC Genomics, 2014, 15, 1025. | |
| Chen R Y, Song W Q, Liang G L. Chromosome atlas of Chinese fruit trees and their close wild relatives. Beijing:Internatinal Academic Publishers, 1993, 1, 351- 353. | |
| Cheng Z K, Buell C R, Wing R A, et al. Toward a cytological characterization of the rice genome. Genome Research, 2002, 11 (12): 2133- 2141. | |
| Chung K S, Weber J A, Hipp A L. 2011. Dynamics of chromosome number and genome size variation in a cytogenetically variable sedge (Carex scoparia var. scoparia, Cyperaceae). American Journal of Botany, 98(1): 122−129. | |
|
Dong F, Song J, Naess S K, et al. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theoretical and Applied Genetics, 2000, 101 (7): 1001- 1007.
doi: 10.1007/s001220051573 |
|
|
Du P, Cui C H, Liu H, et al. Development of an oligonucleotide dye solution facilitates high throughput and cost-efficient chromosome identification in peanut. Plant Methods, 2019, 15, 69.
doi: 10.1186/s13007-019-0451-7 |
|
|
Du P, Li L A, Zhang Z X, et al. Chromosome painting of telomeric repeats reveals new evidence for genome evolution in peanut. Journal of Integrative Agriculture, 2016, 15 (11): 2488- 2496.
doi: 10.1016/S2095-3119(16)61423-5 |
|
|
Du P, Li L N, Liu H, et al. High-resolution chromosome painting with repetitive and single-copy oligonucleotides in Arachis species identifies structural rearrangements and genome differentiation . BMC Plant Biology, 2018, 18 (1): 240.
doi: 10.1186/s12870-018-1468-1 |
|
|
Du P, Zhuang L F, Wang Y Z, et al. Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes . Genome, 2017, 60 (2): 93- 103.
doi: 10.1139/gen-2016-0095 |
|
|
HanY H, Zhang Z H, Thammapichai P, et al. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics, 2015, 200 (3): 771- 779.
doi: 10.1534/genetics.115.177642 |
|
|
He Q Y, Cai Z X, Hu T H, et al. Repetitive sequence analysis and karyotyping reveals centromere-associated DNA sequences in radish (Raphanus sativus L.) . BMC Plant Biology, 2015, 15, 105.
doi: 10.1186/s12870-015-0480-y |
|
| Huang Y J, Xiao L H, Zhang Z, et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition . Gigascience, 2019, 8 (5): 1- 17. | |
|
Jiang J M. Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Research, 2019, 27 (3): 153- 165.
doi: 10.1007/s10577-019-09607-z |
|
| Liu X Y, Sun S, Wu Y, et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species . Plant Journal, 2019, 101 (1): 112- 121. | |
|
Manos P S, Stone D E. Evolution, phylogeny, and systematics of the Juglandaceae. Annals of the Missouri Botanical Garden, 2001, 88 (2): 231- 269.
doi: 10.2307/2666226 |
|
| Mondin M, Aguiar-Perecin, M L. Heterochromatin patterns and ribosomal DNA loci distribution in diploid and polyploid Crotalaria species (Leguminosae, Papilionoideae) and inferences on karyotype evolution . Genome, 2011, 54 (9): 718- 726. | |
|
Moraes A P, Simões A O, Alayon D I O, et al. Detecting mechanisms of karyotype evolution in Heterotaxis (Orchidaceae) . PLoS One, 2016, 11, e0165960.
doi: 10.1371/journal.pone.0165960 |
|
| Roa F, Guerra M. 2012. Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evolutionary Biology, 12: 225. | |
|
Rocha L C, de Oliveira Bustamante F, Silveira R A, et al. Functional repetitive sequences and fragile sites in chromosomes of Lolium perenne L . Protoplasma, 2015, 252 (2): 451- 60.
doi: 10.1007/s00709-014-0690-4 |
|
| Vasconcelos E V, Vasconcelos S, Ribeiro T, et al. Karyotype heterogeneity in Philodendron s. l. (Araceae) revealed by chromosome mapping of rDNA loci . PLoS One, 2018, 13 (11): 1- 15. | |
|
Xin H Y, Zhang T, Wu Y F, et al. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting . Plant Journal, 2020, 101 (2): 253- 264.
doi: 10.1111/tpj.14536 |
|
|
Yakovlev S S, Godelle B, Zoldos V, et al. Evolutionary implications of heterochromatin and rDNA in chromosome number and genome size changes during dysploidy: a case study in Reichardia genus . PLoS One, 2017, 12 (8): e0182318.
doi: 10.1371/journal.pone.0182318 |
|
| Yu F, Zhao X W, Chai J, et al. Chromosome-specific painting unveils chromosomal fusions and distinct allopolyploid species in the Saccharum complex . New Phytologist, 2021, 233 (4): 1953- 1965. | |
|
Zhang J B, Li R Q, Xiang X G, et al. Integrated fossil and molecular data reveal the biogeographic diversification of the Eastern Asian-Eastern North American disjunct hickory genus (Carya Nutt.) . PLoS One, 2013, 8 (7): e70449.
doi: 10.1371/journal.pone.0070449 |
| [1] | Chuanmei Xu,Zihan Wang,Feng Xie,Xuyin Jin,Xinyan Wu,Rong Chen,Jianqin Huang. Comparison of 45S rDNA and 5S rDNA Distribution Characteristics of 12 Bamboo Species [J]. Scientia Silvae Sinicae, 2022, 58(7): 93-102. |
| [2] | Yanmin Li,Deyi Yuan,Tianwen Ye,Ya Chen,Chunxia Han,Shixin Xiao. Karyotype Analysis of 18 Excellent Individuals in F1 Generation of Interspecific Hybridization of Oil-Tea (Camellia oleifera) [J]. Scientia Silvae Sinicae, 2022, 58(4): 165-174. |
| [3] | Lüji Wang,Xiaoxiao Zhang,Jun Wang. Chromosome Behaviors Tracking during Meiosis of Microsporocytes of Allotriploid Populus alba × P. berolinensis 'Yinzhong' Based on 45S rDNA-FISH [J]. Scientia Silvae Sinicae, 2022, 58(3): 69-78. |
| [4] | Tianwen Ye,Deyi Yuan,Yanmin Li,Shixin Xiao,Shoufu Gong,Jian Zhang,Sufang Li,Jian Luo. Ploidy Identification of Camellia hainanica [J]. Scientia Silvae Sinicae, 2021, 57(7): 61-69. |
| [5] | Ruihong Guo,Shuai Qiu,Guangxin Liu,Kai Gao,Jianfen Wei,Mengli Xi. Optimization of Chromosome Preparation and rDNA Physical Localization of Hydrangea macrophylla [J]. Scientia Silvae Sinicae, 2021, 57(11): 59-67. |
| [6] | Xu Zilong, Chen Yicun, Gao Ming, Wu Liwen, Zhao Yunxiao, Wang Yangdong. Research Progress in Sex Differentiation in Angiosperms [J]. Scientia Silvae Sinicae, 2019, 55(8): 157-169. |
| [7] | Tian Mengdi, Li Yanjie, Zhang Pingdong, Wang Jian, Hao Jingyi. Pollen Chromosome Doubling Induced by High Temperature Exposure to Produce Hybrid Triploids in Populus canescens [J]. Scientia Silvae Sinicae, 2018, 54(3): 39-47. |
| [8] | Xu Chuanmei, Huang Jianqin, Wang Zhengjia, Xia Guohua, Zhang Qixiang, Huang Youjun, Zhang Shougong. Meiosis of Pollen Mother Cell and Karyotype of Carya cathayensis [J]. Scientia Silvae Sinicae, 2017, 53(11): 77-84. |
| [9] | Jia Fangxin, Zhou Mingbing, Chen Rong, Yang Haiyun, Gao Peijun, Xu Chuanmei. Karyotype and Genome Size in Four Bamboo Species [J]. Scientia Silvae Sinicae, 2016, 52(9): 57-66. |
| [10] | Li Tiezhu;Tian Dalun;Wuyun Tana;Tan Xiaofeng;Xiang Wenhua;Yan Wende;. Identification and Variation of Tetraploid Camellia oleifera [J]. Scientia Silvae Sinicae, 2009, 12(3): 150-154. |
| [11] | Wang Xiaorong;Tang Haoru;Fu Huaqing;Zhong Bifeng;Xia Wufeng;Tang Haidong;Deng Qunxian. Karyotypes of 15 Introduced Bramble Cultivars (Rubus) [J]. Scientia Silvae Sinicae, 2008, 44(12): 147-150. |
| [12] | Xu Jin;Shi Jisen;Yang Liwei;Wang Guifeng. Cell Biological Characteristics and Abnormal Behavior during the Meiosis of Pollen Mother Cells in Cunninghamia lanceolata [J]. Scientia Silvae Sinicae, 2007, 43(11): 32-36. |
| [13] | Yin Aihua;Jin Hui;Han Zhengmin;Han Sufen. Study on the Relationship between Chromosome Numbers and Nodulation of 18 Species of Leguminous Trees [J]. Scientia Silvae Sinicae, 2006, 42(1): 26-28. |
| [14] | Yumei Wang,Maoxue Li,Ruilin You. OBSERVATION ON CHROMOSOMES OF GINKGO BILOBA BY USING MATERIALS RATHER THAN ROOT-TIP CELLS [J]. Scientia Silvae Sinicae, 1999, 35(5): 2-4. |
| [15] | Xiangyang Kang,Zhiti Zhu,Huibin Lin. STUDY ON THE EFFECTIVE TREATING PERIOD FOR POLLEN CHROMOSOME DOUBLING OF POPULUS TOMENTOSA×P. BOLLEANA [J]. Scientia Silvae Sinicae, 1999, 35(4): 21-24. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||