

Scientia Silvae Sinicae ›› 2022, Vol. 58 ›› Issue (3): 48-58.doi: 10.11707/j.1001-7488.20220306
Previous Articles Next Articles
Zhiyang Li,Xiaolin Chen,Lili Li,Shiping Xu,Yuanhao He*
Received:2021-01-15
Online:2022-03-25
Published:2022-06-02
Contact:
Yuanhao He
CLC Number:
Zhiyang Li,Xiaolin Chen,Lili Li,Shiping Xu,Yuanhao He. Transcriptome Analysis of Camellia olefolia Root and the Endophytic Bacteria Bacillus Subtilis at the Early Stage of Their Interaction[J]. Scientia Silvae Sinicae, 2022, 58(3): 48-58.
Table 1
Primers used for qRT-PCR gene expression analysis"
| 基因名称 Gene name | 引物 Primer(5′-3′) | 扩增长度 Amplification length/bp | 退火温度 Annealing temperature(Tm)/℃ |
| POD | TCATACATTCGGACGGGCTG AGTTGTAAAGGCGTGGGGTC | 104 | 56.9 |
| RBOHC | ATCGCTCAACCATGCCAAGA ATCGCTCAACCATGCCAAGA | 109 | 57.0 |
| TCH4 | AACTCAAGCTTGTGCCTGGA TCATGTGTTGGCCCTTGTGA | 84 | 56.8 |
| TIFY | TTCCCGGCTGATAAGGCAAA TTTTGGACCGGTGTTGAAGC | 105 | 56.3 |
| XTH | GGGGAATTTGAGTGGTGATCCT TGGAAATCAGCAGTCGGGTC | 104 | 57.0 |
| EF1α | CAAAGAAGGGTGCCAAGTGA ACCAAACAACCGACCTACGA | 128 | 56.2 |
Table 2
Summary of De novo data generated for C. oleifera transcriptomes inoculated with endophytic bacteria at different time points"
| 时间 Time/hpi | 总序列数 Total reads | 唯一比对读段 Number of uniquely mapped reads | 比对率 Proportion of mapped reads (%) | GC含量 GC percentage(%) | Q30含量 Q30 percentage(%) |
| 0 | 139 674 846 | 80 575 830 | 69.58 | 43.69 | 92.2 |
| 6 | 130 079 280 | 78 164 416 | 72.83 | 43.89 | 92.19 |
| 12 | 124 726 484 | 72 206 074 | 70.34 | 44.21 | 92.18 |
| 24 | 135 478 312 | 79 027 352 | 70.85 | 43.98 | 92.29 |
| N50长度 N50 length/bp | 1 175 866 | ||||
| 平均长度 Average length/bp | 1 175 866 | ||||
| 比对率 Unique mapped ratio (%) | 71.13 | ||||
| 注释的基因数量 Annotated gene number | 67 700 | ||||

Fig.4
Heat map diagram of the expression levels of DEGs A: Heat map diagram of the expression levels of DEGs which are involved in plant hormone signal transduction. B: Heat map diagram of the expression levels of DEGs which are involved in plant-pathogen interaction. C: Heat map diagram of theexpression levels of DEGs which are involved in plant phenylpropanoid biosynthesis."
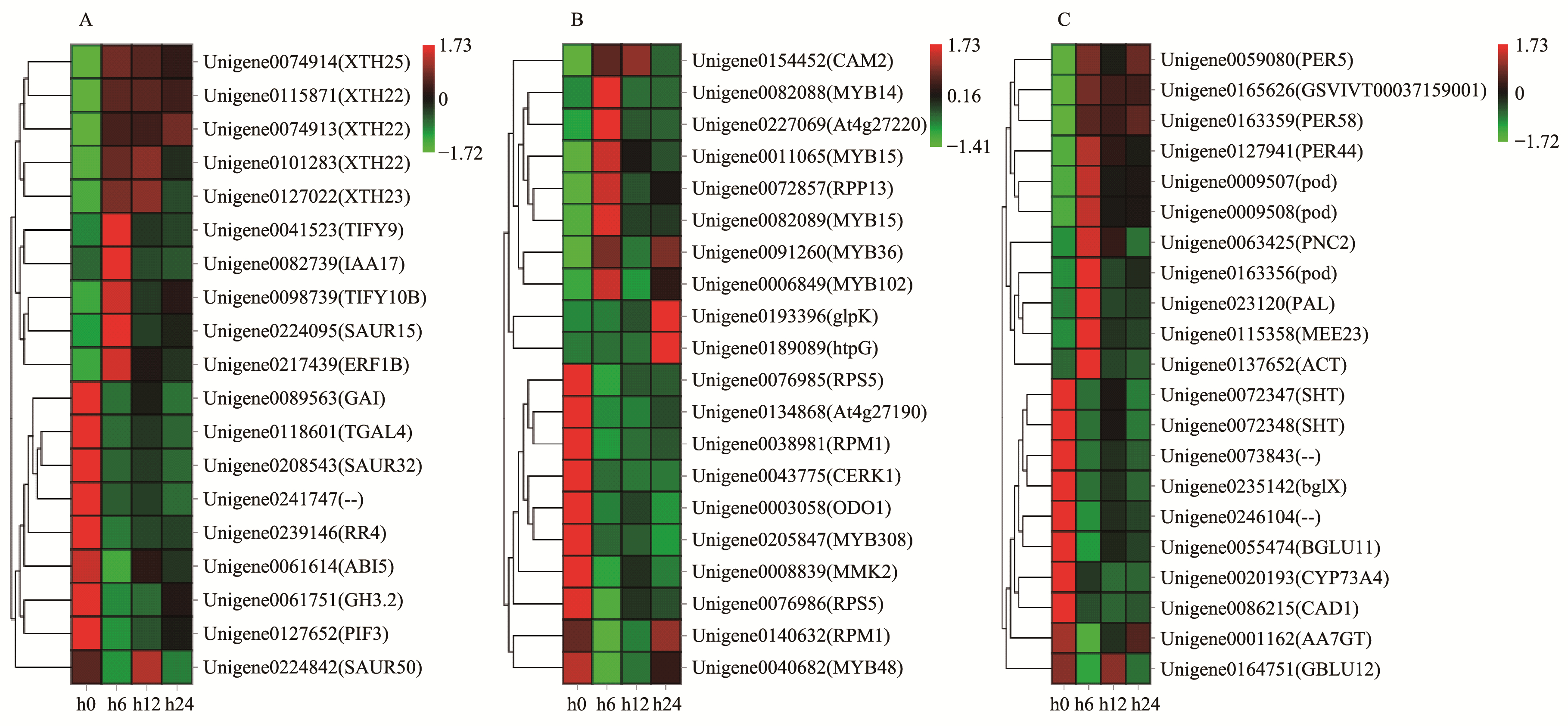
Table 3
DEGs of plant hormone signal transduction pathway"
| 家族 Family | 基因ID Gene ID | 表达量变化的log2值 log2 value of fold change | 描述(符号) Description(Symbol) | ||
| h6 | h12 | h24 | |||
| 生长素响应基因家族SAUR The auxin response gene family SAUR | Unigene0224095 | 5.271 017 | -1.797 32 | 0.397 943 | 生长素诱导蛋白 Auxin-induced protein X15(SAUR15) |
| Unigene0208543 | -2.391 3 | 0.664 469 | -0.731 89 | 生长素诱导蛋白 Auxin-induced protein 15A(SAUR32) | |
| Unigene0224842 | -4.887 15 | 5.342 368 | -3.498 13 | 生长素诱导蛋白 Auxin-induced protein 15A (SAUR50) | |
| 糖苷水解酶家族GH16 Glycoside hydrolase family GH16 | Unigene0074914 | 4.234 786 | -0.121 78 | -0.231 35 | 木葡聚糖内糖基转移酶/水解蛋白酶 Xyloglucan endotransglucosylase/hydrolase protein 23(XTH25) |
| Unigene0115871 | 3.840 573 | -0.004 68 | -0.151 34 | 木葡聚糖内糖基转移酶/水解蛋白酶 Xyloglucan endotransglucosylase/hydrolase protein 22(XTH22) | |
| TIFY家族基因 TIFY family gene | Unigene0041523 | 4.115 512 | -1.814 64 | -0.207 01 | 茉莉酮酸酯ZIM结构域蛋白 Jasmonate ZIM domain-containing protein (TIFY9) |
| Unigene0098739 | 3.869 918 | -1.349 38 | 0.522 57 | 茉莉酮酸酯ZIM结构域蛋白 Jasmonate-zim-domain protein (TIFY10B) | |
| 生长素早期响应基因家族AUX/IAA Auxin early response gene family AUX/IAA | Unigene0082739 | 2.984 088 | -2.344 58 | -0.231 92 | 生长素 Auxin-responsive protein IAA(IAA17) |
| 转录因子bZIP Transcription factor bZIP | Unigene0061614 | -2.504 95 | 1.864 647 | -0.385 94 | 脱落酸应答元件结合因子 ABA responsive element binding factor(ABI5) |
| Unigene0118601 | -4.000 41 | 1.824 728 | -1.405 45 | 含碱性亮氨酸拉链结构域的蛋白 Basic-leucine zipper domain-containing protein(TGAL4) | |
| GH3基因家族 GH3 gene family | Unigene0061751 | -3.476 34 | 1.027 397 | 1.369 855 | 生长素响应GH3基因家族 Auxin responsive GH3 gene family (GH3) |
| DELLA基因家族 DELLA gene family | Unigene0089563 | -7.258 48 | 5.469 323 | -10.347 1 | DELLA蛋白 DELLA protein (GAI) |
Table 4
DEGs of plant-pathogen interaction pathway"
| 家族 Family | 基因ID Gene ID | 表达量变化的log2值 log2 value of fold change | 描述(符号) Description(Symbol) | ||
| h6 | h12 | h24 | |||
| CAM | Unigene0154452 | 6.431 758 | 0.194 622 | -1.359 71 | 钙调蛋白 Calmodulin (CAM2) |
| MYB | Unigene0006849 | 5.059 191 | -3.267 78 | 2.442 811 | MYB相关蛋白Myb4 Myb-related protein Myb4 (MYB102) |
| Unigene0091260 | 3.342 609 | -1.393 12 | 1.400 094 | 转录因子MYB36 Transcription factor MYB36 (MYB36) | |
| Unigene0079478 | 2.615 85 | -2.153 92 | 1.069 583 | 转录因子MYB Transcription factor MYB (MYB4) | |
| RPM | Unigene0140632 | -3.087 74 | 1.679 16 | 1.575 752 | 抗病蛋白 Disease resistance protein (RPM1) |
| Unigene0076986 | -2.419 19 | 1.434 07 | -0.188 74 | 抗病蛋白 Disease resistance protein At4g27220 (RPS5) | |
| Unigene0134868 | -3.151 73 | 0.296 655 | 1.008 148 | 核苷酸结合位点-富含亮氨酸重复蛋白 Nucleotide-binding site leucine-rich repeat protein (At4g27190) | |
| RBOH | Unigene0041782 | 2.166 077 | 0.104 336 | -1.046 16 | 呼吸爆发氧化酶 Respiratory burst oxidase (RBOHC) |
| MAPK | Unigene0176521 | 1.873 724 | -0.689 07 | 0.553 341 | MAPK激酶3 Mitogen-activated protein kinase 3 (NTF4) |
| Unigene0008839 | -1.662 43 | 0.789 679 | -0.486 96 | MAPK同源物 Mitogen-activated protein kinase homolog MMK2(MMK2) | |
| RPM1互作蛋白 RPM1 interacting protein | Unigene0126538 | 1.872 688 | -0.307 55 | -0.702 38 | RPM1互作蛋白 RPM1-interacting protein (RIN4) |
| LysM受体类激酶 LysM receptor kinases | Unigene0043775 | -3.029 47 | -0.579 95 | -0.056 76 | LysM受体类激酶 LysM domain receptor-like kinase (CERK1) |
| 气味结合蛋白 Odorant binding protein | Unigene0003058 | -2.260 62 | 0.780 021 | -1.123 88 | 气味结合蛋白 Protein ODORANT1 (ODO1) |
Table 5
DEGs of phenylpropanoid biosynthesis pathway"
| 家族 Family | 基因ID Gene ID | 表达量变化的log2值 log2 value of fold change | 描述(符号) Description(Symbol) | ||
| h6 | h12 | h24 | |||
| 过氧化物酶基因家族 Peroxidase gene family(POD) | Unigene0059080 | 3.777 526 | -0.643 98 | 0.573 304 | 类木质素形成阴离子过氧化物酶 Lignin-forming anionic peroxidase-like (PER5) |
| Unigene0163356 | 3.407 953 | -1.814 69 | 0.370 634 | 木质素形成阴离子过氧化物酶 Lignin-forming anionic peroxidase (pod) | |
| Unigene0163359 | 1.999 275 | -0.126 23 | 0.149 761 | 类过氧化物酶 Peroxidase A2-like(PER58) | |
| 糖苷水解酶家族 Glycoside hydrolase family(GH) | Unigene0164751 | -1.465 3 | 1.472 489 | -1.055 36 | 类β-葡萄糖苷酶12 Beta-glucosidase 12-like(BGLU12) |
| Unigene0001162 | -1.559 93 | 0.931 72 | 0.399 258 | 类β-葡萄糖苷酶11 Beta-glucosidase 11-like(AA7GT) | |
| Unigene0055474 | -2.519 19 | 1.392 39 | -0.277 75 | β-葡萄糖苷酶 Beta glucosidase 11(BGLU11) | |
| 肉桂醇脱氢酶 Cinnamyl alcohol (CAD) dehydrogenase | Unigene0086215 | -1.826 91 | -0.280 1 | 0.181 386 | 肉桂醇脱氢酶1 Cinnamyl alcohol dehydrogenase 1 (CAD1) |
| 段龙飞, 慕小倩, 李文燕. 茉莉酸信号途径中转录抑制因子JAZ蛋白家族的分子进化分析. 植物学报, 2013, 48 (6): 623- 634. | |
| Duan L F , Mu X Q , Li W Y . Molecular evolution analysis of the JAZ protein family of transcriptional suppressor in jasmonic acid signaling pathway. Acta Botanica Sinica, 2013, 48 (6): 623- 634. | |
| 张佳琦, 胡恒康, 徐川梅, 等. 核桃JrGA2ox基因的克隆、亚细胞定位及功能验证. 林业科学, 2019, 55 (2): 50- 60. | |
| Zhang J Q , Hu H K , Xu C M , et al. Cloning, subcellular mapping and functional verification of JrGa2Ox gene in Walnut. Scientia Silvae Sinicae, 2019, 55 (2): 50- 60. | |
| 王恒煦, 刘泽平, 王志刚, 等. 3株芽孢杆菌在水稻根际定殖促生及其在土壤中的存活. 生态与农村环境学报, 2019, 35 (7): 892- 899. | |
| Wang H X , Liu Z P , Wang Z G , et al. 3 strains of Bacillus colonize and promote growth in rice rhizosphere and their survival in soil. Journal of Ecology and Rural Environment, 2019, 35 (7): 892- 899. | |
|
Banerjee A , Roychoudhury A . WRKY proteins: signaling and regulation of expression during abiotic stress responses. The Scientific World Journal, 2015, 225 (2): 807560- 807561.
doi: 10.1155/2015/807560 |
|
|
Bechinger C . Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science, 1999, 285 (5435): 1896- 1899.
doi: 10.1126/science.285.5435.1896 |
|
|
Bulgarelli D , Rott M , Schlaeppi K , et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature, 2012, 488 (7409): 91- 95.
doi: 10.1038/nature11336 |
|
| Dominik K G , Eric van der G , Thomas R . Phytoalexin transgenics in crop protection-fairy tale with a happy end?. Plant Science, 2012, 195 (94): 54- 70. | |
|
Drogue B , Sanguin H , Chamam A , et al. Plant root transcriptome profiling reveals a strain-dependent response during Azospirillum-rice cooperation. Frontiers in Plant Science, 2014, 5 (10): 607- 608.
doi: 10.3389/fpls.2014.00607 |
|
| Galambos N , Compant S , Moretto M , et al. Humic acid enhances the growth of tomato promoted by endophytic bacterial strains through the activation of Hormone-, Growth-, and Transcription-Related processes. Frontiers in Plant Science, 2020, 11 (21): 1437- 1438. | |
|
Irizarry I , White J . Bacillus amyloliquefaciens alters gene expression, ROS production, and lignin synthesis in cotton seedling roots. Journal of Applied Microbiology, 2018, 124 (6): 1589- 1603.
doi: 10.1111/jam.13744 |
|
|
Ishihama N , Yoshioka H . Post-translational regulation of WRKY transcription factors in plant immunity. Current Opinion in Plant Biology, 2012, 15 (4): 431- 437.
doi: 10.1016/j.pbi.2012.02.003 |
|
|
Johnson C , Arias B J . Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell, 2003, 15 (8): 1846- 1858.
doi: 10.1105/tpc.012211 |
|
|
King E , Wallner A , Rimbault I , et al. Monitoring of rice transcriptional responses to contrasted colonizing patterns of Phytobeneficial burkholderia s. l. reveals a temporal shift in JA systemic response. Frontiers in Plant Science, 2019, 10 (3): 1141- 1141.
doi: 10.3389/fpls.2019.01141 |
|
|
Langmead B , Salzberg S L . Fast gapped-read alignment with Bowtie 2. Nature Methods, 2012, 10 (4): 357- 358.
doi: 10.1038/nmeth.1923 |
|
|
Macho A P , Zipfel C . Plant PRRs and the activation of innate immune signaling. Molecular Cell, 2014, 54 (2): 263- 272.
doi: 10.1016/j.molcel.2014.03.028 |
|
| Naoumkina M A , Zhao Q , Gallego-giraldo L , et al. Genome-wide analysis of phenylpropanoid defence pathways. Molecular Plant Pathology, 2010, 11 (6): 829- 846. | |
|
Pandey S P , Smssich I E . The role of WRKY transcription factors in plant immunity. Plant Physiology, 2009, 150 (4): 1648- 1655.
doi: 10.1104/pp.109.138990 |
|
| Pankievicz V C , Camilios-Neto D , Bonato P , et al. RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Molecular Biology, 2016, 90 (11): 589- 603. | |
|
Pertea G , Huang X , Liang F , et al. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics, 2003, 19 (5): 651- 652.
doi: 10.1093/bioinformatics/btg034 |
|
|
Pinski A , Betekhtin A , Hupert-Kocurek K , et al. Defining the fenetic basis of plant-endophytic bacteria interactions. International Journal of Molecular Sciences, 2019, 28 (4): 1947- 1947.
doi: 10.3390/ijms20081947 |
|
|
Saenz-Mata J , Berenice Salazar-Badillo F , Francisco Jimenez-Bremont J , et al. Transcriptional regulation of Arabidopsis Thaliana WRKY fenes under interaction with beneficial fungus Trichoderma atroviride. Acta Physiologia Plantarum, 2014, 36 (5): 1085- 1093.
doi: 10.1007/s11738-013-1483-7 |
|
|
Sheibani-Tezerji R , Rattei T , Sessitsch A , et al. Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. MBIO, 2015, 6 (21): 14- 15.
doi: 10.1128/mBio.00621-15 |
|
| Shidore T , Dinse T , Ohrlein J , et al. Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environmental Microbiology, 2012, 14 (5): 2775- 2787. | |
|
Singh N , Pandey G K . Calcium signatures and signal transduction schemes during microbe interactions in Arabidopsis Thaliana. Journal of Plant Biochemistry and Biotechnology, 2020, 29 (4): 675- 686.
doi: 10.1007/s13562-020-00604-6 |
|
|
Straub D , Yang H , Liu Y , et al. Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte Herbaspirillum frisingense GSF30T. Journal of Experimental Botany, 2013, 64 (14): 4603- 4615.
doi: 10.1093/jxb/ert276 |
|
|
Stringlis I A , Yu K , Feussner K , et al. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115 (22): E5213- E5222.
doi: 10.1073/pnas.1722335115 |
|
|
Tian W , Hou C , Ren Z , et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature, 2019, 572 (7767): 131- 135.
doi: 10.1038/s41586-019-1413-y |
|
|
Vogt T . Phenylpropanoid biosynthesis. Molecular Plant, 2010, 3 (2): 20- 25.
doi: 10.1093/mp/ssp106 |
|
|
Xu J X , Li Z Y , Lv X , et al. Isolation and characterization of Bacillus Subtilis strain 1-L-29, an endophytic bacteria from Camellia oleifera with antimicrobial activity and efficient plant-root colonization. PLoS ONE, 2020, 15 (4): e0232096.
doi: 10.1371/journal.pone.0232096 |
|
| Yi Y , De Jong A , Frenzel E , et al. Comparative transcriptomics of Bacillus mycoides strains in response to potato-root exudates reveals different genetic adaptation of endophytic and soil isolates. Front Microbiol, 2017, 8 (13): 1487. | |
|
Yimer H Z , Nahar K , Kyndt T , et al. Gibberellin antagonizes jasmonate-induced defense against Meloidogyne graminicola in Rice. New Phytologist, 2018, 218 (2): 646- 660.
doi: 10.1111/nph.15046 |
|
| Yuan P , Jauregui E , Du L , et al. Calcium signatures and signaling events orchestrate plant-microbe interactions. Current Opinion in Plant Biology, 2017, 38 (1): 173- 183. |
| [1] | Xingzhou Chen,Guoying Zhou,Xinggang Chen,Lingyu Jiang,Anhua Bao,Jun Liu. Screening of Effectors of Colletotrichum fructicola in Camellia oleifera [J]. Scientia Silvae Sinicae, 2021, 57(9): 110-120. |
| [2] | Xiya Li,Shengpei Zhang,He Li. Function of Vacuolar Protein Sorting CfVps26 in Colletotrichum fructicola on Camellia oleifera [J]. Scientia Silvae Sinicae, 2021, 57(8): 94-101. |
| [3] | Baicheng Xie,Lingyao Guo,Dongsheng Du,Yan Tan,Guodong Wang. Responses of Camellia oleifera Yield to Heat Accumulation Temperature and High Temperature Days in Key Growth Period [J]. Scientia Silvae Sinicae, 2021, 57(5): 34-42. |
| [4] | Yalan Gao,Yuanhao He,He Li. Biological Function bZIP-Type Transcription Factor CfAp1 in Colletotrichum fructicola [J]. Scientia Silvae Sinicae, 2020, 56(9): 30-39. |
| [5] | Yongli Li,Yahong Wang,Le Chang,Hairu Yu,Zhou Zhou,Wenxia Zhao,Liangjian Qu. Isolation and Identification of Endophytic Bacteria from Xinjiang Wild Apple (Malus sieversii) and Their Antagonism against Three Pathogens of Apple Trees [J]. Scientia Silvae Sinicae, 2020, 56(5): 97-104. |
| [6] | Shouke Zhang,Linxin Fang,Yi Wang,Wei Zhang,Jinping Shu,Yangdong Wang,Haojie Wang. Evaluation Model for Resistance of Camellia oleifera to Curculio chinensis (Coleoptera: Curculionidae) Based on Fruit Properties [J]. Scientia Silvae Sinicae, 2020, 56(12): 67-74. |
| [7] | Li He, Li Sizheng, Wang Yuechen, Liu Jun, Xu Jianping, Zhou Guoying. Identification of the Pathogens Causing Anthracnose of Camellia oleifera in Nursery and Their Resistence to Fungicides [J]. Scientia Silvae Sinicae, 2019, 55(5): 85-94. |
| [8] | Jin Qin, Zhu Danxue, Zhou Guoying, Li He, He Yuanhao, Zhang Qian. Colonization of GFP-Tagged Bacillus subtilis Y13UV in Camellia oleifera [J]. Scientia Silvae Sinicae, 2017, 53(7): 111-117. |
| [9] | Li He, Li Yang, Xu Jianping, Zhou Guoying. Population Genetic Structure of Colletotrichum fructicola from Oil-Tea and Other Host Plants in Hainan province [J]. Scientia Silvae Sinicae, 2016, 52(10): 80-88. |
| [10] | Wang Tao;Li Yong;Piao Chungen;Wang Laifa;Zhu Tianhui. Diversity and Seasonal Fluctuations of Culturable Endophytic Bacteria Isolated from Two Poplars [J]. , 2013, 49(5): 135-146. |
| [11] | Li Jiyuan;Xiao Qing;Li Xinlei;Li Jinming;Lu Yizeng. Soil,Water and Nutrient Loss in Young Plantation of the Inter-Cropped Tea Oil (Camellia oleifera) with Different Crops [J]. Scientia Silvae Sinicae, 2008, 44(4): 167-172. |
| [12] | Wang Xiangnan;Chen Yongzhong;Peng Shaofeng;Yang Xiaohu. Five Elite Varieties of Camellia oleifera [J]. Scientia Silvae Sinicae, 2008, 44(4): 173-174. |
| [13] | Tan Xiaofeng;Chen Hongpeng;Zhang Dangquan;Zeng Yanling;Li Wei;Jiang Yao;Xie Lushan;Hu Xiaoyi;Hu Fangming. Cloning of Full-Length cDNA of FAD2 Gene from Camellia oleifera [J]. Scientia Silvae Sinicae, 2008, 44(3): 70-75. |
| [14] | Han Ninglin. THE TEST FOR COMPARING CAMELLIA OLEIFERA CLONES PLANTING THE NURSE SEED GRAFTINGS [J]. , 1989, 25(4): 375-381. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||