

林业科学 ›› 2020, Vol. 56 ›› Issue (1): 38-53.doi: 10.11707/j.1001-7488.20200105
陆小雨1,2,陈竹1,唐菲1,傅松玲2,任杰1,*
收稿日期:2019-02-01
出版日期:2020-01-25
发布日期:2020-02-24
通讯作者:
任杰
基金资助:Xiaoyu Lu1,2,Zhu Chen1,Fei Tang1,Songling Fu2,Jie Ren1,*
Received:2019-02-01
Online:2020-01-25
Published:2020-02-24
Contact:
Jie Ren
摘要:
目的: 红花槭秋彩叶的形成,与叶片中花青素苷的含量密切相关。本文旨在揭示红花槭中花青素苷的生物合成机理,为其叶色的定向改良提供理论依据。方法: 为解析花青素代谢物积累和基因表达水平的变化,以转色期同时具有绿叶、红叶和黄叶的红花槭特殊单株为材料,用超高效液相色谱串联质谱和高通量RNA测序的方法分别进行代谢组和转录组分析。结果: 1) 在红叶-绿叶、黄叶-绿叶、红叶-黄叶3个比较组中,代谢组正离子模式下分别检测出1 377、1 793、1 098个差异积累代谢物,负离子模式下分别检测出789、699、677个差异积累代谢物:红叶与绿叶相比,矢车菊素苷衍生物、天竺葵素苷元和飞燕草素苷元及其衍生物含量大幅上升;黄叶与绿叶相比,矢车菊素苷衍生物、飞燕草素苷及其衍生物含量增加,而天竺葵素苷及其衍生物减少。2)3个比较组中,转录组测序分别检测出28 536、43 017、27 110个差异表达基因:红叶与绿叶相比,花青素苷合成通路中89.5%的基因表达量增加;黄叶与绿叶相比,花青素苷合成通路中66.7%的基因表达量增加。3)红花槭花青素苷的生物合成中,有29个差异积累的相关代谢物和48个差异表达基因。4)差异代谢物和基因的网络互作分析显示,CHS2, CHS7, CHS8, F3H1, F3H5, F3H7, F3H8, F3H10, F3′H2, LAR, FLS1, FLS2, UFGT4逆向调控天竺葵素苷衍生物和飞燕草素苷衍生物的合成,UFGT5正向调控矢车菊素苷衍生物的合成。结论: 当红花槭叶片秋季变色时,花青素苷通路中大量基因的表达量上调,同时矢车菊素-3-(6″-乙酰半乳糖苷)和矢车菊素-3-阿拉伯糖苷含量大幅上升,此为红花槭叶片变色的主要驱动因子。
中图分类号:
陆小雨,陈竹,唐菲,傅松玲,任杰. 转录组与代谢组联合解析红花槭叶片中花青素苷变化机制[J]. 林业科学, 2020, 56(1): 38-53.
Xiaoyu Lu,Zhu Chen,Fei Tang,Songling Fu,Jie Ren. Combined Transcriptomic and Metabolomic Analysis Reveals Mechanism of Anthocyanin Changes in Red Maple(Acer rubrum) Leaves[J]. Scientia Silvae Sinicae, 2020, 56(1): 38-53.
表1
花青素苷衍生物名称"
| 代码Code | 中文名称Chinese name | 英文名称English name |
| Pg-H-M-C-R-G | 天竺葵素-O(羟基-甲氧基-鼠李糖基-吡喃葡糖苷)-O-吡喃葡糖苷 | Pelargonidin-O-(hydroxy-methoxy-cinnamoyl-L-rhamnopyranosyl-glucopyranoside)-O-glucopyranoside |
| Pg-G-H-G | 天竺葵素-O-(吡喃葡糖基-羟基-肉桂酸基-吡喃葡糖苷)-O-吡喃葡糖苷 | Pelargonidin-O-(glucopyranosyl-hydroxycinnamoyl-glucopyranoside)-O-glucopyranoside |
| Pg-Gsy | 天竺葵素-3-(2-谷氨基-葡糖基芸香糖苷) | Pelargonidin 3-(2-glu-glucosylrutinoside) |
| Pg-Ga | 天竺葵素-5-半乳糖苷 | Pelargonidin 5-galactoside |
| Cy-Acet | 矢车菊素-3-(4-乙酰葡糖苷) | Cyanidin 3-(4-acetylglucoside) |
| Cy-A-Ga | 矢车菊素-3-(6″-乙酰半乳糖苷) | Cyanidin 3-(6″-acetyl-galactoside) |
| Cy-X-H-G | 矢车菊素-O-(吡喃木糖基-羟基肉桂基-吡喃葡糖苷) | Cyanidin-O-(xylopyranosyl-hydroxycinnamoyl-glucopyranosyl) |
| Cy-S | 矢车菊素-3-桑布双糖苷 | Cyanidin 3-sambubioside |
| Cy-Arab | 矢车菊素-3-阿拉伯糖苷 | Cyanidin 3-arabinoside |
| Cy-L | 矢车菊素-3-昆布双糖苷 | Cyanidin 3-laminaribioside |
| Del-M-Glu | 飞燕草素-3-(6″-丙二葡糖苷) | Delphinidin 3-(6″-malonyl-glucoside) |
| Del-Acet | 飞燕草素-3-乙酰葡糖苷 | Delphinidin 3-acetylglucoside |
| Del-di-M | 飞燕草素-3, 5-二-(6-O-丙二葡糖苷) | Delphinidin 3, 5-di(6-O-malonylglucoside) |
| Del-S | 飞燕草素-3-桑布双糖苷 | Delphinidin 3-sambubioside |
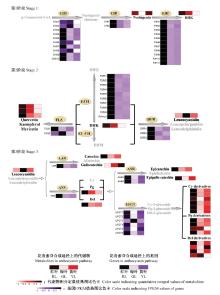
图4
红叶、绿叶和黄叶中花青素苷合成途径上相关基因和代谢物的细节展示 CHS: 查尔酮合成酶;CHI: 查尔酮异构酶;F3H: 黄烷酮3-羟化酶;F3′H: 黄烷酮3′-羟化酶;F3′5′H: 黄烷酮3′5′-羟化酶;DFR: 二氢黄酮醇4-还原酶;ANS: 花青素苷合成酶;UFGT: 类黄酮3-O-葡糖基转移酶;FLS: 黄酮醇合成酶;LAR: 原花青素苷元还原酶;ANR: 花青素苷还原酶。P-Coumaroyl-CoA:对肉桂酸辅酶A;Naringenin chalcone:柚皮素查尔酮;Naringenin:柚皮素;DHK:二氢山奈酚;DHQ:二氢槲皮素;DHM:二氢杨梅素;Quercetin:槲皮素;Kaempferol:山奈酚;Myricetin:杨梅素;Leucocyanidin:原矢车菊素苷元;Leucopelargonidin:原天竺葵素苷元;Leucodelphnidin:原飞燕草素苷元;Catechin:儿茶素;Afzelechin:阿福豆素;Gallocatechin:没食子儿茶素;Cy: 矢车菊素苷;Pg: 天竺葵素苷;Del: 飞燕草素苷;Epicatechin:表儿茶素;Epiafzelechin:表阿福豆素;Epigallocatechin:表没食子儿茶素;Cy-3-glucoside:矢车菊素-3-葡糖苷;Pg-3-glucoside:天竺葵素-3-葡糖苷;Del-3-glucoside:飞燕草素-3-葡糖苷。"
表2
不同比较组间显著富集KEGG通路(修正P≤0.01)"
| 编号No. | 通路名称Pathway | 通路ID Pathway ID | 通路注释的差异表达基因DEGs with pathway annotation | 通路注释的所有基因All genes with pathway annotation | P | 修正P值Corrected P |
| 红叶-绿叶Red leaves-green leaves | ||||||
| 1 | 核糖体Ribosome | ko03010 | 286 | 441 | 2.67E-23 | 3.26E-21 |
| 2 | 卟啉和叶绿素代谢Porphyrin and chlorophyll metabolism | ko00860 | 120 | 193 | 7.66E-10 | 4.67E-08 |
| 3 | 类黄酮生物合成Flavonoid biosynthesis | ko00941 | 76 | 102 | 4.37E-09 | 1.78E-07 |
| 4 | 光合作用-天线蛋白质Photosynthesis-antenna proteins | ko00196 | 58 | 76 | 1.58E-07 | 4.82E-06 |
| 5 | 精氨酸和脯氨酸代谢Arginine and proline metabolism | ko00330 | 171 | 377 | 5.79E-06 | 0.000 137 722 |
| 6 | 半胱氨酸和蛋氨酸代谢Cysteine and methionine metabolism | ko00270 | 224 | 525 | 6.77E-06 | 0.000 137 722 |
| 7 | 苯丙氨酸代谢Phenylalanine metabolism | ko00360 | 115 | 231 | 8.66E-06 | 0.000 150 948 |
| 8 | 精氨酸生物合成Arginine biosynthesis | ko00220 | 86 | 162 | 1.94E-05 | 0.000 295 595 |
| 9 | 甘氨酸、丝氨酸和苏氨酸代谢Glycine, serine and threonine metabolism | ko00260 | 142 | 312 | 2.97E-05 | 0.000 403 202 |
| 10 | 乙醛酸和二羧酸代谢Glyoxylate and dicarboxylate metabolism | ko00630 | 246 | 612 | 6.04E-05 | 0.000 736 680 |
| 11 | 抗坏血酸代谢Ascorbate and aldarate metabolism | ko00053 | 124 | 274 | 0.000 109 140 | 0.001 210 459 |
| 12 | 泛醌和其他萜类化合物-醌生物合成Ubiquinone and other terpenoid-quinone biosynthesis | ko00130 | 88 | 182 | 0.000 194 958 | 0.001 982 071 |
| 13 | 磷酸戊糖途径Pentose phosphate pathway | ko00030 | 151 | 360 | 0.000 349 958 | 0.003 284 225 |
| 14 | 苯丙氨酸、酪氨酸和色氨酸生物合成Phenylalanine, tyrosine and tryptophan biosynthesis | ko00400 | 87 | 186 | 0.000 479 922 | 0.004 182 176 |
| 15 | 植物-病原体相互作用Plant-pathogen interaction | ko04626 | 230 | 595 | 0.000 584 893 | 0.004 757 129 |
| 16 | 光合生物中的碳固定Carbon fixation in photosynthetic organisms | ko00710 | 229 | 593 | 0.000 625 676 | 0.004 770 781 |
| 17 | 谷胱甘肽代谢Glutathione metabolism | ko00480 | 158 | 388 | 0.000 727 198 | 0.005 094 233 |
| 18 | 苯丙烷类生物合成Phenylpropanoid biosynthesis | ko00940 | 96 | 214 | 0.000 751 608 | 0.005 094 233 |
| 19 | 氰氨基酸代谢Cyanoamino acid metabolism | ko00460 | 50 | 94 | 0.000 929 480 | 0.005 968 242 |
| 20 | 泛酸和CoA生物合成Pantothenate and CoA biosynthesis | ko00770 | 65 | 133 | 0.000 998 095 | 0.006 088 377 |
| 21 | 类胡萝卜素生物合成Carotenoid biosynthesis | ko00906 | 67 | 140 | 0.001 276 839 | 0.007 417 825 |
| 22 | 缬氨酸、亮氨酸和异亮氨酸的生物合成Valine, leucine and isoleucine biosynthesis | ko00290 | 55 | 110 | 0.001 581 266 | 0.008 768 837 |
| 23 | 果糖和甘露糖代谢Fructose and mannose metabolism | ko00051 | 125 | 304 | 0.001 850 311 | 0.009 814 695 |
| 24 | 二苯乙烯类、二芳基庚烷类和姜辣素类的生物合成Stilbenoid, diarylheptanoid and gingerol biosynthesis | ko00945 | 27 | 42 | 0.001 955 262 | 0.009 939 251 |
| 绿叶-黄叶Green leaves-yellow leaves | ||||||
| 1 | 核糖体Ribosome | ko03010 | 322 | 441 | 1.91E-13 | 2.33E-11 |
| 2 | 卟啉和叶绿素代谢Porphyrin and chlorophyll metabolism | ko00860 | 136 | 193 | 5.56E-06 | 0.000 339 316 |
| 红叶-黄叶Red leaves-yellow leaves | ||||||
| 1 | 类黄酮生物合成Flavonoid biosynthesis | ko00941 | 73 | 102 | 1.82E-10 | 2.22E-08 |
| 2 | 苯丙烷类生物合成Phenylpropanoid biosynthesis | ko00940 | 108 | 214 | 3.92E-08 | 2.39E-06 |
| 3 | 甘油脂代谢Glycerolipid metabolism | ko00561 | 202 | 520 | 1.16E-06 | 4.73E-05 |
| 4 | 调节自噬Regulation of autophagy | ko04140 | 118 | 287 | 2.67E-05 | 0.000 812 852 |
| 5 | 核糖体Ribosome | ko03010 | 164 | 441 | 6.85E-05 | 0.001 671 012 |
| 6 | 淀粉和蔗糖代谢Starch and sucrose metabolism | ko00500 | 370 | 1 149 | 0.000 168 590 | 0.003 427 987 |
| 7 | 苯丙氨酸、酪氨酸和色氨酸生物合成Phenylalanine, tyrosine and tryptophan biosynthesis | ko00400 | 78 | 186 | 0.000 350 292 | 0.005 966 480 |
| 8 | 糖胺聚糖降解Glycosaminoglycan degradation | ko00531 | 23 | 33 | 0.000 391 245 | 0.005 966 480 |
| 9 | N-聚糖生物合成N-glycan biosynthesis | ko00510 | 75 | 182 | 0.000 664 722 | 0.009 010 676 |
| 10 | 果糖和甘露糖代谢Fructose and mannose metabolism | ko00051 | 113 | 304 | 0.000 842 426 | 0.009 555 408 |
| 11 | 萜类骨架生物合成Terpenoid backbone biosynthesis | ko00900 | 67 | 160 | 0.000 896 156 | 0.009 555 408 |
| 12 | 脂肪酸延伸率Fatty acid elongation | ko00062 | 25 | 41 | 0.000 939 876 | 0.009 555 408 |
| 冯立娟, 苑兆和, 尹燕雷, 等. 美国红枫叶色表达期间相关物质的研究. 山东林业科, 2008, (4): 9- 11. | |
| Feng L J , Yuan Z H , Yin Y L , et al. Studies on the related substances for color expression in American maple during the leaf color transition. Journal of Shandong Forestry Science & Technology, 2008, (4): 9- 11. | |
| 李玲. 2013. 五种红色叶植物叶片色素分析. 西安: 西北农林科技大学硕士学位论文. | |
| Li L . The research on the chromogenic pigment of leaves in five red plants. Xi'an: MS thesis of Northwest A&F University, 2013. | |
| 任杰, 丁增成, 唐菲, 等. 加拿大红枫的引种及繁育技术研究. 中国农学通报, 2013, 29 (1): 37- 41. | |
| Ren J , Ding Z C , Tang F , et al. Study on introduction and propagation techniques of red maple(Acer rubrum L. ). Chinese Agricultural Science Bulletin, 2013, 29 (1): 37- 41. | |
| 石柏林, 吴家森, 钟泰林. 6种槭树属植物种子特性及其发芽试验. 浙江林业科技, 2006, 26 (3): 38- 40. | |
| Shi B L , Wu J S , Zhong T L . Research on seed properties and germination test of six species of Acer. Journal of Zhejiang Forestry Science & Technology, 2006, 26 (3): 38- 40. | |
| 夏涛, 高丽萍. 类黄酮及茶儿茶素生物合成途径及其调控研究进展. 中国农业科学, 2009, 42 (8): 2899- 2908. | |
| Xia T , Gao L P . Advances in biosynthesis pathways and regulation of flavonoids and catechins. Scientia Agricultura Sinica, 2009, 42 (8): 2899- 2908. | |
| Abouzaid M M , Helson B V , Nozzolillo C , et al. Ethyl m-digallate from red maple, Acer rubrum L. , as the major resistance factor to forest tent caterpillar, Malacosoma disstria Hbn. Journal of Chemical Ecology, 2001, 27 (12): 2517- 2527. | |
|
Abrams M D . The red maple paradox. Bioscience, 1998, 48 (5): 355- 364.
doi: 10.2307/1313374 |
|
|
Alexander H D , Arthur M A . Implications of a predicted shift from upland oaks to red maple on forest hydrology and nutrient availability. Canadian Journal of Forest Research, 2010, 40 (4): 716- 726.
doi: 10.1139/X10-029 |
|
|
Cai Y , Weng K , Guo Y , et al. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing. Metabolomics, 2015, 11 (6): 1575- 1586.
doi: 10.1007/s11306-015-0809-4 |
|
|
Chen Z , Lu X Y , Xuan Y , et al. Transcriptome analysis based on a combination of sequencing platforms provides insights into leaf pigmentation in Acer rubrum. BMC Plant Biology, 2019, 19 (1): 240- 256.
doi: 10.1186/s12870-019-1850-7 |
|
|
Cho K , Cho K S , Sohn H B , et al. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. Journal of Experimental Botany, 2016, 67 (5): 1519.
doi: 10.1093/jxb/erv549 |
|
| Chu Y X , Chen H R , Wu A Z , et al. Expression analysis of dihydroflavonol 4-reductase genes in Petunia hybrida. Genetics & Molecular Research, 2015, 14 (2): 5010- 5021. | |
|
Cole T , Williams B A , Geo P , et al. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nature Biotechnology, 2010, 28 (5): 511- 515.
doi: 10.1038/nbt.1621 |
|
|
Davies K M , Schwinn K E , Deroles S C , et al. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica, 2003, 131 (3): 259- 268.
doi: 10.1023/A:1024018729349 |
|
|
Deng X , Bashandy H , Ainasoja M , et al. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytologist, 2014, 201 (4): 1469- 1483.
doi: 10.1111/nph.12610 |
|
| Deshmukh R , Sonah H , Patil G , et al. Integrating omic approaches for abiotic stress tolerance in soybean. Frontiers in Plant Science, 2014, 5, 244. | |
|
Dunn W B , Broadhurst D , Begley P , et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 2011, 6 (7): 1060- 1083.
doi: 10.1038/nprot.2011.335 |
|
|
Ford C M , Boss P K , Hoj P B . Cloning and characterization of Vitis vinifera UDP-glucose: Flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. Journal of Biological Chemistry, 1998, 273 (15): 9224- 9233.
doi: 10.1074/jbc.273.15.9224 |
|
|
Geoffroy T R , Fortin Y , Stevanovic T . Process optimisation for pilot-scale production of maple bark extracts, natural sources of antioxidants, phenolics, and carbohydrates. Chemical Papers, 2018, 72 (5): 1125- 1137.
doi: 10.1007/s11696-017-0355-9 |
|
|
Guo X , Chao Y , Le L , et al. Transcriptome of the floral transition in Rosa chinensis 'Old Blush'. Bmc Genomics, 2017, 18, 199.
doi: 10.1186/s12864-017-3584-y |
|
|
Han Y , Huang K , Liu Y , et al. Functional analysis of two flavanone-3-hydroxylase genes from Camellia sinensis: A critical role in flavonoid accumulation. Genes, 2017, 8 (11): 300.
doi: 10.3390/genes8110300 |
|
|
Hauke J , Kossowski T . Comparison of values of Pearson's and Spearman's correlation coefficients on the same sets of data. Quaestiones Geographicae, 2011, 30 (2): 87- 93.
doi: 10.2478/v10117-011-0021-1 |
|
|
He H , Ke H , Keting H , et al. Flower colour modification of chrysanthemum by suppression of F3'H and overexpression of the exogenous Senecio cruentus F3'5'H gene. PLoS One, 2013, 8 (11): e74395.
doi: 10.1371/journal.pone.0074395 |
|
| He J , Giusti M M . Anthocyanins: natural colorants with health-promoting properties. Annual Review of Food Science & Technology, 2010, 1 (1): 163- 187. | |
|
Hu C , Gong Y , Jin S , et al. Molecular analysis of a UDP-glucose: flavonoid 3-O-glucosyltransferase(UFGT) gene from purple potato (Solanum tuberosum). Molecular Biology Reports, 2011, 38 (1): 561- 567.
doi: 10.1007/s11033-010-0141-z |
|
| Kalubi K N , Mehes-Smith M , Omri A . Comparative analysis of metal translocation in red maple (Acer rubrum) and trembling aspen(Populus tremuloides) populations from stressed ecosystems contaminated with metals. Chemistry & Ecology, 2016, 32 (4): 312- 323. | |
| Kitada C , Gong Z , Tanaka Y , et al. Differential expression of two cytochrome P450s involved in the biosynthesis of flavones and anthocyanins in chemo-varietal forms of Perilla frutescens. Plant & Cell Physiology, 2001, 42 (12): 1338- 1344. | |
|
Kobayashi S , Ishimaru M , Ding C K , et al. Comparison of UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Science, 2001, 160 (3): 543- 550.
doi: 10.1016/S0168-9452(00)00425-8 |
|
|
Koes R , Verweij W , Quattrocchio F . Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science, 2005, 10 (5): 236- 242.
doi: 10.1016/j.tplants.2005.03.002 |
|
|
Koes R E , Spelt C E , Mol J N M . The chalcone synthase multigene family of Petunia hybrida (V30): differential, light-regulated expression during flower development and UV light induction. Plant Molecular Biology, 1989, 12 (2): 213- 225.
doi: 10.1007/BF00020506 |
|
| Kuittinen H , Aguadé M . Nucleotide variation at the chalcone isomerase locus in Arabidopsis thaliana. Genetics, 2000, 155 (2): 863- 872. | |
|
Kumar A , Singh B , Singh K . Functional characterization of flavanone 3-hydroxylase gene from Phyllanthus emblica (L.). Journal of Plant Biochemistry & Biotechnology, 2015, 24 (4): 453- 460.
doi: 10.1007/s13562-014-0296-0 |
|
|
Li B , Dewey C N . RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Bmc Bioinformatics, 2011, 12 (1): 323- 323.
doi: 10.1186/1471-2105-12-323 |
|
| Li L , Liu Y , Liu Y , et al. Physiological response and resistance of three cultivars of Acer rubrum L. to continuous drought stress. Acta Ecologica Sinica, 2015, 35 (6): 196- 202. | |
|
Liu Y , Shi Z , Maximova S , et al. Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biology, 2013, 13 (1): 202.
doi: 10.1186/1471-2229-13-202 |
|
|
Lu X , Chen Z , Gao J , et al. Combined metabolome and transcriptome analyses of photosynthetic pigments in red maple. Plant Physiol Biochem, 2020, 154, 476- 490.
doi: 10.1016/j.plaphy.2020.06.025 |
|
|
Matile P , Hortensteiner S , Thomas H . Chlorophyll degradation. Annual Review of Plant Biology, 2006, 57, 55- 77.
doi: 10.1146/annurev.arplant.57.032905.105212 |
|
|
Moreno-Risueno M A , Busch W , Benfey P N . Omics meet networks—using systems approaches to infer regulatory networks in plants. Current Opinion in Plant Biology, 2010, 13 (2): 126- 131.
doi: 10.1016/j.pbi.2009.11.005 |
|
|
Moschen S , Bengoa Luoni S , Di Rienzo J A , et al. Integrating transcriptomic and metabolomic analysis to understand natural leaf senescence in sunflower. Plant Biotechnol J, 2016, 14 (2): 719- 734.
doi: 10.1111/pbi.12422 |
|
| Nam-Soo K , Min-Ji I , Kabwe N . Determination of DNA methylation associated with Acer rubrum(red maple) adaptation to metals: analysis of global DNA modifications and methylation-sensitive amplified polymorphism. Ecology & Evolution, 2016, 6 (16): 5749- 5760. | |
| Nkongolo K , Theriault G , Michael P . Differential levels of gene expression and molecular mechanisms between red maple (Acer rubrum) genotypes resistant and susceptible to nickel toxicity revealed by transcriptome analysis. Ecology & Evolution, 2018, 8 (10): 4876- 4890. | |
|
Piero A R L , Consoli A , Puglisi I , et al. Anthocyaninless cultivars of sweet orange lack to express the UDP-glucose flavonoid 3-O-glucosyl transferase. Journal of Plant Biochemistry & Biotechnology, 2005, 14 (1): 9- 14.
doi: 10.1007/BF03263217 |
|
|
Qian L , Liu Y , Qi Y , et al. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. Journal of Experimental Botany, 2014, 65 (12): 3157- 3164.
doi: 10.1093/jxb/eru168 |
|
|
Rai A , Saito K , Yamazaki M . Integrated omics analysis of specialized metabolism in medicinal plants. Plant Journal, 2017, 90 (4): 764- 787.
doi: 10.1111/tpj.13485 |
|
|
Rai A , Yamazaki M , Takahashi H , et al. RNA-seq transcriptome analysis of Panax japonicus, and its comparison with other Panax species to identify potential genes involved in the saponins biosynthesis. Frontiers in Plant Science, 2016, 7, 148.
doi: 10.3389/fpls.2016.00481 |
|
|
Rebeki A , Lonari Z , Petrovi S , et al. Pearson's or Spearman's correlation coefficient: Which one to use?. Poljoprivreda, 2015, 21 (2): 47- 54.
doi: 10.18047/poljo.21.2.8 |
|
|
Riañopachón D M , Nagel A , Neigenfind J , et al. The GABI primary database: GABIPD—integration of plant 'omics'-data in gene context. Nature Precedings, 2009,
doi: 10.1038/npre.2008.1684.1 |
|
| Royer M , Diouf P N , Stevanovic T . Polyphenol contents and radical scavenging capacities of red maple (Acer rubrum L. ) extracts. Food & Chemical Toxicology, 2011, 49 (9): 2180- 2188. | |
|
Sapna K , Nie J , Chen H S , et al. Evaluation of gene association methods for coexpression network construction and biological knowledge discovery. PLoS ONE, 2012, 7 (11): e50411.
doi: 10.1371/journal.pone.0050411 |
|
|
Schmitzer V , Osterc G , Veberic R , et al. Correlation between chromaticity values and major anthocyanins in seven Acer palmatum Thunb. cultivars. Scientia Horticulturae, 2009, 119 (4): 442- 446.
doi: 10.1016/j.scienta.2008.09.003 |
|
|
Shen Q , Fu L , Dai F , et al. Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. Bmc Genomics, 2016, 17, 889.
doi: 10.1186/s12864-016-3242-9 |
|
|
Shimada N , Ayabe S I . A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiology, 2003, 131 (3): 941- 951.
doi: 10.1104/pp.004820 |
|
| Sibley J L , Eakes D J , Gilliam C H , et al. Foliage characteristics of selected red maple cultivars. Research Report, 1995, 1 (1): 40- 41. | |
|
Smith C A , Want E J , O'maille G , et al. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry, 2006, 78 (3): 779- 787.
doi: 10.1021/ac051437y |
|
|
Song T T , Xu H H , Sun N , et al. Metabolomic analysis of alfalfa (Medicago sativa L. ) root-symbiotic rhizobia responses under alkali stress.. Frontiers in Plant Science, 2017, 8, 1208.
doi: 10.3389/fpls.2017.0120 |
|
|
Springob K , Nakajima J I , Yamazaki M , et al. Recent advances in the biosynthesis and accumulation of anthocyanins. Natural Product Reports, 2003, 20 (3): 288- 303.
doi: 10.1039/b109542k |
|
|
Tanaka Y , Sasaki N , Ohmiya A . Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant Journal, 2008, 54 (4): 733- 749.
doi: 10.1111/j.1365-313X.2008.03447.x |
|
|
Wang J , Zhang T , Shen X , et al. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics, 2016, 12 (7): 1- 10.
doi: 10.1007/s11306-016-1050-5 |
|
|
Want E J , Wilson I D , Gika H , et al. Global metabolic profiling procedures for urine using UPLC-MS. Nature Protocols, 2010, 5 (6): 1005- 1018.
doi: 10.1038/nprot.2010.50 |
|
|
Williams R S , Lincoln D E , Norby R J . Development of gypsy moth larvae feeding on red maple saplings at elevated CO2 and temperature. Oecologia, 2003, 137 (1): 114- 122.
doi: 10.1007/s00442-003-1327-z |
|
| Wu X , Prior R L . Systematic identification and characterization of anthocyanins by HPLC-ESI-MS/MS in common foods in the United States: fruits and berries. Journal of Agricultural & Food Chemistry, 2005, 53 (7): 2589- 2599. | |
| Zheng Y , Wang C Y , Wang S Y , et al. Effect of high-oxygen atmospheres on blueberry phenolics, anthocyanins, and antioxidant capacity. Journal of Agricultural & Food Chemistry, 2003, 51 (24): 7162- 7169. |
| [1] | 苏佳露,史无双,杨雅运,王星,丁雨龙,林树燕. 6个竹种叶色与光合色素含量及叶片结构比较[J]. 林业科学, 2020, 56(7): 194-203. |
| [2] | 朱沛煌,陈妤,朱灵芝,李荣,季孔庶. 马尾松转录组密码子使用偏好性及其影响因素[J]. 林业科学, 2020, 56(4): 74-81. |
| [3] | 赵清泉, 池玉杰, 张健, 冯连荣. 偏肿革裥菌在木质环境下转录组构建与相关基因表达分析[J]. 林业科学, 2019, 55(8): 95-105. |
| [4] | 韩小红, 卢赐鼎, 华银, 林浩宇, 是雨霏, 吴松青, 张飞萍, 梁光红. 星天牛转录组及三大解毒酶家族相关基因系统发育分析[J]. 林业科学, 2019, 55(5): 104-113. |
| [5] | 刘果, 陈鸿鹏, 吴志华, 彭彦, 谢耀坚. 南美油藤种子发育过程的代谢组学和转录组学联合分析[J]. 林业科学, 2019, 55(5): 169-179. |
| [6] | 黄亚丽,张军,樊英利,刘易超,杨敏生. 遮荫对中华金叶榆和鑫叶栾叶片呈色及相关生理指标的影响[J]. 林业科学, 2019, 55(10): 171-180. |
| [7] | 候亚会, 严善春, 李志强. 对苯二酚对台湾乳白蚁诱食效果及其诱导代谢的响应[J]. 林业科学, 2018, 54(9): 97-103. |
| [8] | 张恩亮, 马玲玲, 杨如同, 李林芳, 汪庆, 李亚, 王鹏. IBA诱导楸树嫩枝扦插不定根发育的转录组分析[J]. 林业科学, 2018, 54(5): 48-61. |
| [9] | 张芹, 徐宗大, 赵凯, 李晓伟, 张罗沙, 张启翔. 梅花花青素苷调控基因PmMYB1的分离及功能分析[J]. 林业科学, 2018, 54(10): 64-72. |
| [10] | 曹亚兵, 翟晓巧, 邓敏捷, 赵振利, 范国强. 泡桐丛枝病发生与代谢组变化的关系[J]. 林业科学, 2017, 53(6): 85-93. |
| [11] | 毛伟兵, 陈发菊, 王长兰, 梁宏伟. 楸树雄性不育花芽转录组测序及分析[J]. 林业科学, 2017, 53(6): 141-150. |
| [12] | 唐海霞, 杜淑辉, 邢世岩, 桑亚林, 李际红, 刘晓静, 孙立民. 银杏性别决定相关基因的筛选[J]. 林业科学, 2017, 53(2): 76-82. |
| [13] | 丁昌俊, 张伟溪, 高暝, 黄秦军, 褚延广, 苏晓华. 不同生长势美洲黑杨转录组差异分析[J]. 林业科学, 2016, 52(3): 47-58. |
| [14] | 时小东, 朱学慧, 盛玉珍, 庄国庆, 陈放. 基于转录组序列的楠木SSR分子标记开发[J]. 林业科学, 2016, 52(11): 71-78. |
| [15] | 张敏, 黄利斌, 周鹏, 钱猛, 窦全琴. 榉树秋季转色期叶色变化的生理生化[J]. 林业科学, 2015, 51(8): 44-51. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||