

Scientia Silvae Sinicae ›› 2020, Vol. 56 ›› Issue (1): 120-132.doi: 10.11707/j.1001-7488.20200112
• Articles • Previous Articles Next Articles
Wei Jin1,2,Yuzhan Yang2,Haijing Sun2,Lianqing Chen2,Zhilin Yuan1,2,*
Received:2018-10-17
Online:2020-01-25
Published:2020-02-24
Contact:
Zhilin Yuan
Supported by:CLC Number:
Wei Jin,Yuzhan Yang,Haijing Sun,Lianqing Chen,Zhilin Yuan. Diversity of Ectomycorrhizal Fungi a Seed Collecting Forest of Quercus virginiana[J]. Scientia Silvae Sinicae, 2020, 56(1): 120-132.
Table 1
Information of ectomycorrhizal root tips samples used for pyrosequencing"
| 采样时间 Sampling time | 外生菌根颜色 Ectomycorrhizal color | 样品编号 Sample No. | 测序样品数 Number of samples for pyrosequencing |
| 2017-12-26 | 黄白Yellow & white | FLSYYW0 | 3 |
| 黑Black | FLSYB0 | 2 | |
| 2018-04-17 | 白White | FLSYW1 | 6 |
| 黄Yellow | FLSYY1 | 1 | |
| 黑Black | FLSYB1 | 12 | |
| 2018-05-29 | 白White | FLSYW2 | 3 |
| 黄Yellow | FLSYY2 | 3 | |
| 黑Black | FLSYB2 | 3 | |
| 深红Carmine | FLSYR2 | 3 | |
| 乳白色Oyster white | FLSYC2 | 3 | |
| 2018-08-06 | 白White | FLSYW3 | 3 |
| 黄Yellow | FLSYY3 | 3 | |
| 黑Black | FLSYB3 | 3 |
Table 2
Morphological descriptions of Q. virginiana-associated ectomycorrhizas and the corresponding fungal identities"
| 外形结构External structures | 解剖结构Anatomic structures | 外生菌根真菌种类鉴定 | |||||
| 菌根颜色 Color | 分支方式 Type of branching | 表面性状 Surface characters | 外层菌套 Outer layer ofmantle | 内层菌套 Inner layer of mantle | 外延菌丝 Emanating mycelium | 根状菌索 Rhizomorph | |
| 白色 White | 不规则羽状分支 Irregularly pinnate | 密羊毛状 Densely woolly | A型 Type A | 密丝组织 Plectenchymatous | 多 Much | 未分化 Undifferentiated | Scleroderma |
| 黑色 Black | 羽状或塔状 Pinnate or pyramid | 光滑 Smooth | L型 Type L | 拟薄壁组织 Pseudoparenchymatous | 少 Less | 未分化 Undifferentiated | Tomentella |
| 黄色 Yellow | 不规则羽状分支偶见塔状 Irregularly pinnate, pyramid occasionally | 短刺状 Short spiny | D型 Type D | 密丝组织 Plectenchymatous | 少 Less | 未分化 Undifferentiated | Russula |
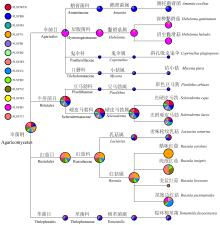
Fig.6
Relative abundance of ectomycorrhizal fungal communities at different taxonomic levels in Q. virginiana The relative abundance is expressed as a percentage. The size of pie chart indicates the relative abundance of corresponding taxonomic level in samples. OTUs not assigned to species level are not included for analysis. "

|
陈益泰, 陈雨春, 黄一青, 等. 抗风耐盐常绿树种弗吉尼亚栎引种初步研究. 林业科学研究, 2007. 20 (4): 542- 546.
doi: 10.3321/j.issn:1001-1498.2007.04.020 |
|
|
Chen Y T , Chen Y C , Huang Y Q , et al. Preliminary study on Quercus virginiana introduction in Eastern China. Forest Research, 2007. 20 (4): 542- 546.
doi: 10.3321/j.issn:1001-1498.2007.04.020 |
|
|
陈益泰, 王树凤, 陈雨春, 等. 弗吉尼亚栎种子产量、脱落过程与种子形态特征的变异及稳定性. 林业科学研究, 2015. 28 (4): 524- 530.
doi: 10.3969/j.issn.1001-1498.2015.04.011 |
|
|
Chen Y T , Wang S F , Chen Y C , et al. Variation and stability of seed yield, fall-off process, seed size and form in live oak. Forest Research, 2015. 28 (4): 524- 530.
doi: 10.3969/j.issn.1001-1498.2015.04.011 |
|
| 陈益泰, 王树凤, 孙海菁, 等. 弗吉尼亚栎引种研究与应用. 浙江: 浙江科学技术出版社. 2017. | |
| Chen Y T , Wang S F , Sun H J , et al. The introduction and applications of Quercus virginiana. Zhejiang: Zhejiang Science Publishing House. 2017. | |
|
陈益泰, 孙海菁, 王树凤, 等. 5种北美栎树在我国长三角地区的引种表现. 林业科学研究, 2013. 26 (3): 344- 351.
doi: 10.3969/j.issn.1001-1498.2013.03.013 |
|
|
Chen Y T , Sun H J , Wang S F , et al. Growth performances of five north Ameirican oak species in Yangzi River Delta of China. Forest Research, 2013. 26 (3): 344- 351.
doi: 10.3969/j.issn.1001-1498.2013.03.013 |
|
| 陈雨春,王杰,宋文君,等. 2007.硝酸银和生根粉在弗吉尼亚栎扦插繁殖中的应用.林业科技开发, 21(3):97-98. | |
| Chen Y C, Wang J, Song W J, et al. 2007. China Forestry Science and Technology, 21(3): 97-98.[in Chinese]. | |
| 陈云, 马克明. 城市菌根真菌多样性、变化机制及功能应用. 生态学报, 2016. 36 (14): 4221- 4232. | |
| Chen Y , Ma K M . Mycorrhizal fungi in urban environments:diversity, mechanism, and application. Acta Ecologica Sinica, 2016. 36 (14): 4221- 4232. | |
| 王树凤, 孙海菁, 陈益泰, 等. 模拟干旱胁迫下弗吉尼亚栎苗木叶片相关生理参数的分析. 南京林业大学学报:自然科学版, 2011. 35 (6): 6- 10. | |
| Wang S F , Sun H J , Chen Y T , et al. Analysis of physiological indexes of Quercus virginiana under drought stress. Journal of Nanjing Forestry University:Natural Science Edition, 2011. 35 (6): 6- 10. | |
| 王 树凤 , 胡 韵雪 , 李 志兰 , et al. 盐胁迫对弗吉尼亚栎生长及矿质离子吸收、运输和分配的影响. 生态学报, 2010. 30 (17): 4609- 4616. | |
| Wang S F, Hu Y X, Li Z L, et al. 2010. Effect of NaCl stress on growth and mineral ion uptake, transportation and distribution of Quercus virginiana, 30(17): 4609-4616.[in Chinese]. | |
| Agerer R. 2012. Colour atlas of ectomycorrhizae, 15th ed. Einhorn-Verlag, Schwäbisch Gmünd. | |
|
Bokulich N A , Sathish S , Faith J J , et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature methods, 2013. 10 (1): 57- 59.
doi: 10.1038/nmeth.2276 |
|
|
Caporaso J G , Kuczynski J , Stombaugh J , et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods, 2010. 7 (5): 335- 336.
doi: 10.1038/nmeth.f.303 |
|
| Castaño C , Alday , J G , Parladé J , et al. Seasonal dynamics of the ectomycorrhizal fungus Lactarius vinosus are altered by changes in soil moisture and temperature. Soil Biology & Biochemistry, 2017. 115, 253- 260. | |
|
Cavender-Bares J , Izzo A , Robinson R , et al. Changes in ectomycorrhizal community structure on two containerized oak hosts across an experimental hydrologic gradient. Mycorrhiza, 2009. 19 (3): 133- 142.
doi: 10.1007/s00572-008-0220-3 |
|
| Cairney J W G, Chambers S M. 1999. Ectomycorrhizal fungi: key genera in profile. Springer Berlin Heidelberg. | |
| Davies Jr F T , Call C A . Mycorrhizae, survival and growth of selected woody plant species in lignite overburden in Texas. Agriculture, Ecosystems & Environment, 1990. 31 (3): 243- 252. | |
|
Edgar R C . UPARSE:highly accurate OTU sequences from microbial amplicon reads. Nature methods, 2013. 10 (10): 996- 998.
doi: 10.1038/nmeth.2604 |
|
|
Guevara G , Bonito G , Trappe J M , et al. New north American truffles (Tuber spp.) and their ectomycorrhizal associations. Mycologia, 2013. 105 (1): 194- 209.
doi: 10.3852/12-087 |
|
|
Haas B J , Gevers D , Earl A M , et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Research, 2011. 21 (3): 494- 504.
doi: 10.1101/gr.112730.110 |
|
|
Hazard C , Johnson D . Does genotypic and species diversity of mycorrhizal plants and fungi affect ecosystem function?. New Phytologist, 2018. 220 (4): 1122- 1128.
doi: 10.1111/nph.15010 |
|
| Howe V K. 1964. Mycorrhizal fungi of white oak. Ames: PhD thesis of Iowa State University of Science and Technology. | |
|
Ihrmark K , Bodeker I T M , Cruzmartinez K , et al. New primers to amplify the fungal ITS2 region evaluation by 454-sequencing of artificial and natural communities. FMES Microbiology Ecology, 2012. 82 (3): 666- 677.
doi: 10.1111/j.1574-6941.2012.01437.x |
|
|
Jiao S , Liu Z S , Lin Y B , et al. Bacterial communities in oil contaminated soils:biogeography and co-occurrence patterns. Soil Biology and Biochemistry, 2016. 98, 64- 73.
doi: 10.1016/j.soilbio.2016.04.005 |
|
| Kim C S , Nam J W , Jo J W , et al. Studies on seasonal dynamics of soil-higher fungal communities in Mongolian oak-dominant Gwangneung forest in Korea. Journal of Microbiology, 2016. 54 (1): 14- 22. | |
| Kropp B R, Mueller G M. 1999. Laccaria: key genera in profile. Ectomycorrhizal fungi: key genera in profile. Cairney J W G, Chambers S M (eds). Berlin: Springer Press, 65-88 | |
|
Magoč T , Salzberg S L . FLASH:fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 2011. 27 (21): 2957- 2963.
doi: 10.1093/bioinformatics/btr507 |
|
| Marx D H , Murphy M , Parrish T , et al. Root response of mature live oaks in coastal South Carolina to root zone inoculations with ectomycorrhizal fungal inoculants. Journal of Arboriculture, 1997. 23 (6): 257- 263. | |
|
Morris M H , Pérez-Pérez M A , Smith M E , et al. Multiple species of ectomycorrhizal fungi are frequently detected on individual oak root tips in a tropical cloud forest. Mycorrhiza, 2008. 18 (8): 375- 383.
doi: 10.1007/s00572-008-0186-1 |
|
|
Richard F , Roy M , Shahin O , et al. Ectomycorrhizal communities in a Mediterranean forest ecosystem dominated by Quercus ilex:seasonal dynamics and response to drought in the surface organic horizon. Annals of Forest Science, 2011. 68 (1): 57- 68.
doi: 10.1007/s13595-010-0007-5 |
|
| Smith S E , Read D J . Mycorrhizal symbiosis (Second Edition). London: Academic Press. 1997. | |
|
Szuba A . Ectomycorrhiza of Populus. Forest Ecology and Management, 2015. 347, 156- 169.
doi: 10.1016/j.foreco.2015.03.012 |
|
|
Voříšková J , Brabcová V , Cajthaml T , et al. Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytologist, 2014. 201 (1): 269- 278.
doi: 10.1111/nph.12481 |
|
|
Walker J F , Jr M O , Horton J L . Seasonal dynamics of ectomycorrhizal fungus assemblages on oak seedlings in the southeastern Appalachian Mountains. Mycorrhiza, 2008. 18 (3): 123- 132.
doi: 10.1007/s00572-008-0163-8 |
|
| Walker J , Miller O , Horton J . Hyperdiversity of ectomycorrhizal fungus assemblages on oak seedlings in mixed forests in the southern Appalachian Mountains. Molecular Ecology, 2010. 14 (3): 829- 838. | |
|
Warcup J H . Ectomycorrhizal associations of Australian indigenous plants. New Phytologist, 1980. 85 (4): 531- 535.
doi: 10.1111/j.1469-8137.1980.tb00768.x |
|
|
Yamamoto K , Endo N , Degawa Y , et al. First detection of Endogone ectomycorrhizas in natural oak forests. Mycorrhiza, 2017. 27 (3): 295- 301.
doi: 10.1007/s00572-016-0740-1 |
| [1] | Pang Li, Zhou Zhichun, Zhang Yi, Feng Zhongping. Effects of Simulated Continuous N Sedimentation on the Ectomycorrhizal Symbiosis and soil Nutrients of 2-Year-Old Pinus massoniana Seedlings Under Low P Stress [J]. Scientia Silvae Sinicae, 2016, 52(8): 138-145. |
| [2] | Song Fuqiang, Kong Xiangshi, Li Jize, Chang Wei. Screening the Related Genes in the AM Fungi and Amorpha fruticosa Symbiosis with the Suppression Subtractive Hybridization Technique [J]. Scientia Silvae Sinicae, 2014, 50(11): 64-74. |
| [3] | Zhang Zhiguang. Symbiosis Mechanism of Forestry-Paper Integration from the Perspective of Green Supply Chain [J]. Scientia Silvae Sinicae, 2011, 47(2): 111-117. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||