

林业科学 ›› 2021, Vol. 57 ›› Issue (9): 140-151.doi: 10.11707/j.1001-7488.20210914
彭欣1,王瀚棠1,郭春晖1,杨振德1,2,*,周静1,王雪1,丁芷柔1
收稿日期:2020-12-24
出版日期:2021-09-25
发布日期:2021-11-29
通讯作者:
杨振德
基金资助:Xin Peng1,Hantang Wang1,Chunhui Guo1,Zhende Yang1,2,*,Jing Zhou1,Xue Wang1,Zhirou Ding1
Received:2020-12-24
Online:2021-09-25
Published:2021-11-29
Contact:
Zhende Yang
摘要:
目的: 对入侵性致瘿害虫桉树枝瘿姬小蜂(膜翅目:姬小蜂科)进行EST-SSR引物开发和隐种鉴定,为桉树枝瘿姬小蜂种群遗传学研究及其防治提供理论依据。方法: 采用Illumina HiSeqTM 2000测序平台对3个地理种群的桉树枝瘿姬小蜂雌性成虫进行转录组测序,利用MISA和Primer Premier 3软件进行EST-SSR标记的搜索、挖掘和引物设计。选取400对EST-SSR引物,结合国外报道的14对多态性G-SSR引物,应用1%琼脂糖凝胶电泳检测扩增效率,并用8%变性聚丙烯酰胺凝胶电泳验证引物多态性。基于桉树枝瘿姬小蜂COI基因序列数据对我国14个地理种群320个桉树枝瘿姬小蜂雌性成虫样本进行隐种鉴定。结果: 1)共获得277 048 525条clean reads,包括82.86 G个核苷酸,组装后得到44 878个unigene,平均长度1 082.76 bp,N50为1 976 bp;利用MISA软件搜索到EST-SSR位点14 190个,其中主要重复类型是单核苷酸重复(占总EST-SSR的69.63%),其次是二核苷酸重复和三核苷酸重复(分别占总EST-SSR的16.17%、13.51%)。2)400对EST-SSR引物中有205对引物可有效扩增出目的片段,扩增效率51.25%,国外报道的14对G-SSR引物均能扩增出目的片段;3)用8%变性聚丙烯酰胺凝胶电泳验证SSR引物多态性,最终筛选出10对多态性良好的EST-SSR引物,国外报道的14对G-SSR引物中仅有LiSS2、LiSS5、LiSS13在我国桉树枝瘿姬小蜂样本中存在多态性;4)从我国14个桉树枝瘿姬小蜂地理种群中共获得320条605 bp的COI基因序列,基于桉树枝瘿姬小蜂COI基因序列构建的NJ系统发育树表明,我国仅存在A、B 2种类型的桉树枝瘿姬小蜂隐种,样本比例约为1:2。结论: 筛选出10对适合我国桉树枝瘿姬小蜂种群遗传学研究的EST-SSR引物。我国存在A、B 2种类型的桉树枝瘿姬小蜂隐种,其中隐种A首次在我国发现,桉树枝瘿姬小蜂的隐种鉴定可为该害虫在生物防治中采用正确的生物型提供依据。
中图分类号:
彭欣,王瀚棠,郭春晖,杨振德,周静,王雪,丁芷柔. 入侵性致瘿害虫桉树枝瘿姬小蜂(膜翅目: 姬小蜂科)EST-SSR开发及隐种鉴定[J]. 林业科学, 2021, 57(9): 140-151.
Xin Peng,Hantang Wang,Chunhui Guo,Zhende Yang,Jing Zhou,Xue Wang,Zhirou Ding. EST-SSR Development and Cryptic Species Identification of the Invasive Gall-Causing Pest Leptocybe invasa (Hymenopetra: Eulophidae)[J]. Scientia Silvae Sinicae, 2021, 57(9): 140-151.
表1
桉树枝瘿姬小蜂14个地理种群采样信息"
| 种群代码 Population code | 采集地 Sample sites | 经纬度 Longitude and latitude | 海拔 Elevation/m | 采样时间 Sample time |
| SCDY | 四川德阳Deyang, Sichuan | 104°17′E,31°10′N | 512.3 | 2019-12 |
| SCPZH | 四川攀枝花Panzhihua, Sichuan | 101°75′E, 26°49′N | 1 137.2 | 2018-07 |
| JXGZ | 江西赣州Ganzhou, Jiangxi | 114°43′E, 25°36′N | 195.6 | 2016-07 |
| FJSM | 福建三明Sanming, Fujian | 117°57′E, 26°30′N | 177.6 | 2019-09 |
| HNDZ | 海南儋州Danzhou, Hainan | 108°56′E, 18°09′N | 24.6 | 2017-07 |
| YNKM | 云南昆明Kunming, Yunnan | 103°10′E, 26°06′N | 1 846.5 | 2019-07 |
| GXFCG1 | 广西防城港1 Fangchenggang 1, Gaungxi | 108°1′E, 22°14′N | 250.0 | 2019-07 |
| GXFCG2 | 广西防城港2 Fangchenggang 2, Gaungxi | 107°47′E, 21°57′N | 220.0 | 2019-10 |
| GXNN1 | 广西南宁1 Nanning 1, Guangxi | 108°17′E, 22°51′N | 143.3 | 2019-06 |
| GXNN2 | 广西南宁2 Nanning 2, Guangxi | 108°17′E, 22°50′N | 156.4 | 2019-06 |
| GXWZ | 广西梧州Wuzhou, Guangxi | 111°03′E, 23°03′N | 76.2 | 2016-07 |
| GXLB | 广西来宾Laibin, Guangxi | 108°24′E, 23°16′N | 361.3 | 2020-06 |
| GXYL | 广西玉林Yulin, Guangxi | 110°01′E, 22°30′N | 150.5 | 2019-08 |
| GXQZ | 广西钦州Qinzhou, Guangxi | 109°12′E, 21°52′N | 45.3 | 2020-07 |
表2
桉树枝瘿姬小蜂转录组组装质量"
| 项目Item | 转录本 Transcripts | 独立基因 Unigenes | ||||
| 数量Number | 百分比Percentage(%) | 数量Number | 百分比Percentage(%) | |||
| 长度范围 Sequence length | 300~500 bp | 24 604 | 31.95 | 21 646 | 48.23 | |
| 500~1 000 bp | 15 989 | 20.76 | 10 931 | 24.36 | ||
| 1 000~2 000 bp | 14 026 | 18.21 | 6 272 | 13.98 | ||
| >2 000 bp | 22 390 | 29.07 | 6 029 | 13.43 | ||
| 总数Total number | 77 009 | 100.00 | 44 878 | 100.00 | ||
| 总长度Total length/bp | 140 690 637 | — | 48 592 158 | — | ||
| N50长度N50 length/bp | 3 568 | — | 1 976 | — | ||
| 平均长度Mean length/bp | 1 826.94 | — | 1 082.76 | — | ||
表3
桉树枝瘿姬小蜂不同类型EST-SSRs重复次数统计"
| 重复类型 Repeat type | 重复次数Repeated number | 合计 Total | 比例 Ratio(%) | ||||||||
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ≥12 | |||
| 单核苷酸Mononucleotide | — | — | — | — | — | — | 3 526 | 1 867 | 4 488 | 9 881 | 69.63 |
| 二核苷酸Dinucleotide | — | — | 795 | 548 | 426 | 313 | 166 | 45 | 2 | 2 295 | 16.17 |
| 三核苷酸Trinucleotide | — | 1 005 | 632 | 272 | 8 | 0 | 0 | 0 | 0 | 1 917 | 13.51 |
| 四核苷酸Tetranucleotide | 0 | 75 | 10 | 2 | 0 | 0 | 1 | 0 | 0 | 88 | 0.62 |
| 五核苷酸Pentanucleotide | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 5 | 0.04 |
| 六核苷酸Hexanucleotide | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 4 | 0.03 |
| 总计Total | 0 | 1 084 | 1 438 | 824 | 435 | 313 | 1 968 | 981 | 7 147 | 14 190 | 100.00 |
| 比例Ratio(%) | 0 | 7.64 | 10.13 | 5.81 | 3.07 | 2.21 | 13.87 | 6.91 | 50.37 | 100.00 | — |
表4
桉树枝瘿姬小蜂EST-SSRs重复基元类型统计"
| 重复基元类型 Repeat motifs type | 数量(百分比) Number(percentage) | 占总SSR比率 Percentage in total SSR(%) | 重复基元类型 Repeat motifs type | 数量(百分比) Number(percentage) | 占总SSR比率 Percentage in total SSR(%) | |
| 单核苷酸Mononucleotide | 三核苷酸Trinucleotide | |||||
| A/T | 9 686(98.03%) | TGC/GCA | 262(13.67%) | |||
| C/G | 195(1.97%) | CAG/CTG | 240(12.52%) | |||
| 合计Subtotal | 9 881(100%) | 69.63 | AGC/GCT | 192(10.02%) | ||
| 二核苷酸Dinucleotide | TCG/CGA | 104(5.43%) | ||||
| GA/TC | 603(26.27%) | AAT/ATT | 91(4.75%) | |||
| AG/CT | 531(23.14%) | GCG/CGC | 89(4.64%) | |||
| AT/AT | 244(10.63%) | GAC/GTC | 84(4.38%) | |||
| AC/GT | 232(10.11%) | GGC/GCC | 82(4.28%) | |||
| CA/TG | 232(10.11%) | 其他Others | 773(40.31%) | |||
| TA/TA | 218(9.50%) | 合计Subtotal | 1 917(100%) | 13.51 | ||
| GC/GC | 121(5.27%) | 四、五、六核苷酸Tetra-, penta-and hexa-nucleotide | 97(100%) | 0.69 | ||
| CG/CG | 114(4.97%) | |||||
| 合计Subtotal | 2 295(100%) | 16.17 | 总计Total | 14 190(100%) | 100 |
表5
桉树枝瘿姬小蜂10对多态性EST-SSRs与国外3对多态性G-SSRs引物信息①"
| 编号 ID | 引物序列 Primers(5′-3′) | 重复基元 Reapeat motifs | Ta | NA | 等位基因大小 Alleles size/bp | |
| c120771 | F: AGCCAAAAGGGGTTTGTTCT | R: ACTCAGCAACAGGTGTCACG | (AG)6 | 56 | 2 | 248、252 |
| c125483 | F: AGCGTTCAACCGAAAAGAGA | R: TAAGTCGCTCGCTGTGTGTC | (TC)8 | 56 | 4 | 213、215、217、219 |
| c124062 | F: CGTCTGTTCAGTCCTCCTCC | R: CATTGCAAGCTACAGTCCGA | (CG)7 | 58 | 2 | 145、155 |
| c120888 | F: GCGCGCGTCTATATACTTCC | R: TACGCGCACGAGTTGTATGT | (CT)6 | 57 | 2 | 233、239 |
| c121460 | F: CGCTCTCACGAGGAGAGACT | R: ACATCCGCGACAACTTCTCT | (ATA)7 | 58 | 3 | 196、199、217 |
| c121011 | F: TGAAGACGGCGAAGAGAAGT | R: ATACAGGTTACGCGCGAGAG | (CA)9 | 57 | 2 | 123、127 |
| c69914 | F: ATCGAATGGCCGTATTTCAA | R: CTTGCGGAGAAATCAAGGAG | (TA)10 | 54 | 4 | 198、200、202、204 |
| c121749 | F: GTATACGAGGGGGAGGGAAA | R: ACAGTTGCTGCTGTACGTGG | (CAG)6 | 58 | 2 | 249、258 |
| c127471 | F: ATGTCGAAGGGCAATTTCTG | R: ACTCGGAATTCAATCAACGC | (TGT)7 | 56 | 2 | 220、229 |
| c123946 | F: GTGTCGATTGGCGCTATTGT | R: CGTGTGTGAGAGTGCGAAAT | (GCT)5 | 56 | 4 | 237、240、246、249 |
| LiSS2 | F: CCATATTGGGTCCACCTACC | R: ACCGTCCTTGCGTATACAGG | (AC)12 or (AC)20 | 56 | 9 | 189、191、193、211、213、215、217、243、245 |
| LiSS5 | F: TCGTGTTTACCACCTGACCA | R: AGAGTGCTCAGGCTCGACAT | (AGC)9 | 56 | 2 | 351、354 |
| LiSS13 | F: TGGTACAAATCCCGTCTATGG | R: CGCAACGGTACAGAAATTCA | (ACGC)7 | 54 | 2 | 141、149 |
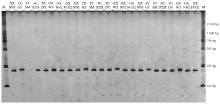
图1
桉树枝瘿姬小蜂c127471引物的变性聚丙烯酰胺凝胶电泳 M:DNA分子量标记DNA molecular weight marker;GXNN1:广西南宁1 Nanning 1, Guangxi;JXGZ:江西赣州Ganzhou, Jiangxi;FJSM:福建三明Sanming, Fujian;SCPZH:四川攀枝花Panzhihua, Sichuan;SCDY:四川德阳Deyang, Sichuan;GXWZ:广西梧州Wuzhou, Guangxi;GXNN2:广西南宁2 Nanning 2, Guangxi;GXFCG2:广西防城港2 Fangchenggang 2, Guangxi."

表6
桉树枝瘿姬小蜂隐种鉴定结果统计"
| 种群代码 Population code | 样本量 Samples | 隐种A数量(比例) Cryptic species A number(Ratio) | 隐种B数量(比例) Cryptic species B number(Ratio) | 隐种C数量(比例) Cryptic species C number(Ratio) |
| SCDY | 32 | 0(0.00%) | 32(100.00%) | 0(0.00%) |
| SCPZH | 32 | 11(34.38%) | 21(65.62%) | 0(0.00%) |
| JXGZ | 32 | 30(93.75%) | 2(6.25%) | 0(0.00%) |
| FJSM | 32 | 0(0.00%) | 32(100.00%) | 0(0.00%) |
| HNDZ | 4 | 0(0.00%) | 4(100.00%) | 0(0.00%) |
| YNKM | 3 | 0(0.00%) | 3(100.00%) | 0(0.00%) |
| GXFCG1 | 32 | 30(93.75%) | 2(6.25%) | 0(0.00%) |
| GXFCG2 | 32 | 1(3.12%) | 31(96.88%) | 0(0.00%) |
| GXNN1 | 32 | 2(6.25%) | 30(93.75%) | 0(0.00%) |
| GXNN2 | 32 | 1(3.12%) | 31(96.88%) | 0(0.00%) |
| GXWZ | 32 | 29(84.38%) | 3(15.62%) | 0(0.00%) |
| GXLB | 16 | 0(0.00%) | 16(100.00%) | 0(0.00%) |
| GXYL | 3 | 0(0.00%) | 3(100.00%) | 0(0.00%) |
| GXQZ | 6 | 0(0.00%) | 6(100.00%) | 0(0.00%) |
| 总计Total | 320 | 104(32.50%) | 216(67.50%) | 0(0.00%) |
表7
全球桉树枝瘿姬小蜂隐种比例统计"
| 国外种群 Foreign population | 样本量 Sample | 隐种数量(比例)Cryptic species (Ratio) | mtDNA COI基因序列的GenBank登录号 GenBank accession numbers for mtDNA COI gene sequences | ||
| A | B | C | |||
| 加纳Ghana | 8 | 0(0.00%) | 8(100.00%) | 0(0.00%) | MH093117—MH093119, MH093168—MH093172 |
| 肯尼亚Kenya | 49 | 49(100.00%) | 0(0.00%) | 0(0.00%) | MH093190—MH093193, MH093226—MH093228, MH093410—MH093426, MH092440—MH093445 MH093446—MH093451, MH093427—MH093439 |
| 莫桑比克 Mozambique | 36 | 36(100.00%) | 0(0.00%) | 0(0.00%) | MH093197—MH093199, MH093234—MH093238, MH093305—MH093309, MH093453—MH094375 |
| 南非 South Africa | 38 | 34(89.47%) | 4(10.53%) | 0(0.00%) | MH093310—MH093324, MH093186, MH093241—MH093243, MH093325—MH093339, MH093111, MH093112, MH093180, MH093181 |
| 突尼斯Tunisia | 3 | 3(100.00%) | 0(0.00%) | 0(0.00%) | KP233982— KP233984 |
| 乌干达Uganda | 23 | 23(100.00%) | 0(0.00%) | 0(0.00%) | MH093194—MH093196, MH093239, MH093240, MH093246, MH093252—MH093259, MH093394—MH093402 |
| 津巴布韦Zimbabwe | 1 | 1(100.00%) | 0(0.00%) | 0(0.00%) | MH093476 |
| 非洲总计 Africa total | 158 | 146(92.41%) | 12(7.59%) | 0(0.00%) | — |
| 中国江西赣州 Ganzhou, Jiangxi,China | 15 | 0(0.00%) | 15(100.00%) | 0(0.00%) | KP233985— KP233988, KP233990—KP233993, JQ289999—JQ280005 |
| 以色列Israel | 33 | 33(100.00%) | 0(0.00%) | 0(0.00%) | MH093187—MH093189, MH093207—MH093210, MH093218—MH093223, MH093260—MH093279 |
| 老挝Laos | 38 | 31(81.58%) | 7(18.42%) | 0(0.00%) | MH093200—MH093204, MH093280—MH093289, MH093452, MH093146, MH093147, MH093149—MH093152, MH093355, MH093148, MH093349—MH093354, MH093345—MH093348, MH093340—MH093343 |
| 马来西亚Malaysia | 9 | 0(0.00%) | 9(100.00%) | 0(0.00%) | MH093113—MH093116, MH093175—MH093179 |
| 越南Vietnam | 10 | 1(10.00%) | 9(90.00%) | 0(0.00%) | MH093139—MH093145, MH093173, MH093174, MH093344 |
| 泰国Thailand | 65 | 31(47.69%) | 34(52.31%) | 0(0.00%) | MH093120—MH093138, MH093153—MH093167, MH093211, MH093212, MH093217, MH093245, MH093247—MH093251, MH093372—MH093393 |
| 亚洲总计 Asia total | 170 | 96(56.47%) | 74(43.53%) | 0(0.00%) | — |
| 澳大利亚Australia | 114 | 0(0.00%) | 109(95.61%) | 5(4.39%) | MH093063, MH093048—MH093052, MH093053, MH093064—MH093074, MH093054, MH093055, MH093182—MH093185, MH093056, MH093075— MH093078, MH093082—MH093086, MH093087, MH093088, MH093057—MH093062, MH093010—MH093042, MH093001—MH093005, MH093006—MH093009, MH093043—MH093047, MH093094, MH093095, MH093079, MH093089— MH093093, MH093080, MH093081, MH093096—MH093110 |
| 大洋洲总计 Australasia total | 114 | 0(0.00%) | 109(95.61%) | 5(4.39%) | — |
| 意大利Italy | 38 | 38(100.00%) | 0(0.00%) | 0(0.00%) | MH093205, MH093206, MH093224, MH093225, MH093290—MH093304, MH093361—MH093371, KP233972— KP233979 |
| 土耳其Turkey | 1 | 1(100.00%) | 0(0.00%) | 0(0.00%) | KP233954 |
| 欧洲总计 Europe total | 39 | 39(100.00%) | 0(0.00%) | 0(0.00%) | — |
| 巴西Brazil | 27 | 27(100.00%) | 0(0.00%) | 0(0.00%) | MH093213—MH093216, MH093229—MH093233, MH093244, MH093403—MH093409, MH093356— MH093360, MH093477—MH093481, |
| 阿根廷Argentina | 3 | 3(100.00%) | 0(0.00%) | 0(0.00%) | KP233979—KP233981 |
| 美洲总计 America total | 30 | 30(100.00%) | 0(0.00%) | 0(0.00%) | — |
| 总计Total | 511 | 311(60.86%) | 195(38.16%) | 5(0.98%) | — |
|
陈华燕, 姚婕敏, 许再福. 桉树枝瘿姬小蜂雄虫在中国的首次发现. 环境昆虫学报, 2009, 31 (3): 285- 287.
doi: 10.3969/j.issn.1674-0858.2009.03.015 |
|
|
Chen H Y , Yao J M , Xu Z F . First description of the male of Leptocybe invasa Fisher & La Salle(Hymenoptera: Eulophidae) from China. Journal of Environmental Entomology, 2009, 31 (3): 285- 287.
doi: 10.3969/j.issn.1674-0858.2009.03.015 |
|
| 郭欢, 王刚, 张树田, 等. 基于RNA-seq数据的窄足真蚋SSR分子标记开发. 昆虫学报, 2018, 61 (7): 815- 824. | |
| Guo H , Wang G , Zhang S T , et al. Development of SSR primers for Simulium(Eusimulium) angustipes (Diptera: Simuliidae) based on RNA-seq dataset. Acta Entomologica Sinica, 2018, 61 (7): 815- 824. | |
| 郭睿, 陈华枝, 庄天艺, 等. 利用转录组数据开发意大利蜜蜂的SSR分子标记. 安徽农业大学学报, 2018, 45 (3): 404- 408. | |
| Guo R , Chen H Z , Zhuang T Y , et al. Exploitation of SSR markers for Apis mellifera ligustica based on transcriptome data. Journal of Anhui Agricultural University, 2018, 45 (3): 404- 408. | |
| 韩小红, 王伊凡, 卢赐鼎, 等. 星天牛转录组SSR位点特征分析. 应用昆虫学报, 2019, 56 (6): 1299- 1308. | |
| Han X H , Wang Y F , Lu C D , et al. Characteristics of the SSR loci in the Anoplophora chinensis transcriptome. Chinese Journal of Applied Entomology, 2019, 56 (6): 1299- 1308. | |
| 黄梦伊, 赵佳强, 石娟. 基于MaxEnt对桉树枝瘿姬小蜂在中国发生趋势的预测. 北京林业大学学报, 2020, 42 (11): 64- 71. | |
| Huang M Y , Zhao J Q , Shi J . Predicting occurrence tendency of Leptocybe invasa in China based on MaxEnt. Journal of Beijing Forestry University, 2020, 42 (11): 64- 71. | |
| 寇冀蒙. 2020. 伴生细菌在桉树枝瘿姬小蜂克服桉树抗性中的机制研究. 哈尔滨: 东北林业大学硕士学位论文. | |
| Kou J M. 2020. The role of associated bacteria of Leptocybe invasa Fisher & La Salle(Hymenoptera: Eulophidae) in the resistance of its host to Eucalyptus. Harbin: MS thesis of Northeast Forestry University. [in Chinese] | |
| 黎东海, 赵萍. 基于转录组数据的齿缘刺猎蝽微卫星分子标记开发. 昆虫学报, 2019, 62 (6): 694- 702. | |
| Li D H , Zhao P . Development of microsatellite markers based on the transcriptome data of Sclomina erinacea(Heteroptera: Reduviidae). Acta Entomologica Sinica, 2019, 62 (6): 694- 702. | |
| 李微, 张蕾, 程云霞, 等. 应用转录组测序高通量发掘东方粘虫SSR标记. 植物保护学报, 2017, 44 (3): 377- 384. | |
| Li W , Zhang L , Cheng Y X , et al. High-throughput discovery of microsatellite markers based on transcriptome sequencing in the oriental armyworm, Mythimna separata(Walker). Journal of Plant Protection, 2017, 44 (3): 377- 384. | |
| 刘慎思, 张桂芬, 武强, 等. 桔小实蝇幼体及成虫残体DNA条形码识别技术的建立与应用. 昆虫学报, 2012, 55 (3): 336- 343. | |
| Liu S S , Zhang G F , Wu Q , et al. Establishment and application of DNA barcoding technology for identification of the immatures and adult debris of Bactrocera dorsalis(Hendel)(Diptera: Tephritidae). Acta Entomologica Sinica, 2012, 55 (3): 336- 343. | |
|
罗基同, 蒋金培, 王缉健, 等. 广西博白桉树枝瘿姬小蜂生物学特性研究. 中国森林病虫, 2011, 30 (4): 10- 12.
doi: 10.3969/j.issn.1671-0886.2011.04.004 |
|
|
Luo J T , Jiang J P , Wang J J , et al. Bionomics of Leptocybe invasa in Bobai of Guangxi Zhuang Autonomous Region. Forest Pest and Disease, 2011, 30 (4): 10- 12.
doi: 10.3969/j.issn.1671-0886.2011.04.004 |
|
|
钱万强, 万方浩. 中国启动IAS1000计划. 中国农业科学, 2018, 51 (22): 4395- 4396.
doi: 10.3864/j.issn.0578-1752.2018.22.017 |
|
|
Qian W Q , Wan F H . China launches the IAS1000 project. Scientia Agricultura Sinica, 2018, 51 (22): 4395- 4396.
doi: 10.3864/j.issn.0578-1752.2018.22.017 |
|
| 桑迪, 徐叶, 王伟亮, 等. 基于转录组数据的意大利蝗微卫星位点分析与分子标记开发. 应用昆虫学报, 2020, 57 (3): 658- 666. | |
| Sang D , Xu Y , Wang W L , et al. Analysis of micro-satellite loci from Calliptamus italicus(Orthopera: Acrididae) based on a transcriptome dataset. Chinese Journal of Applied Entomology, 2020, 57 (3): 658- 666. | |
| 孙陶泽, 母洪娜, 王良桂, 等. 桂花(Osmanthus fragrans)转录组SSR特征分析. 分子植物育种, 2019, 17 (7): 2258- 2263. | |
| Sun T Z , Mu H N , Wang L G , et al. Characteristics analysis of SSR in the transcriptome of Osmanthus fragrans. Molecular Plant Breeding, 2019, 17 (7): 2258- 2263. | |
| 魏丹丹, 石俊霞, 张夏瑄, 等. 基于转录组数据的桔小实蝇微卫星位点信息分析. 应用生态学报, 2014, 25 (6): 1799- 1805. | |
| Wei D D , Shi J X , Zhang X X , et al. Analysis of microsatellite loci from Bactrocera dorsalis based on transcriptome dataset. Journal of Applied Ecology, 2014, 25 (6): 1799- 1805. | |
|
吴耀军, 蒋学建, 李德伟, 等. 我国发现1种重要的林业外来入侵害虫——桉树枝瘿姬小蜂(膜翅目: 姬小蜂科). 林业科学, 2009, 45 (7): 161- 163.
doi: 10.3321/j.issn:1001-7488.2009.07.028 |
|
|
Wu Y J , Jiang X J , Li D W , et al. Leptocybe invasa, a new invasive forest pest making galls on twigs and leaves of Eucalyptus trees in China(Hymenoptera: Eulophidae). Scientia Silvae Sinicae, 2009, 45 (7): 161- 163.
doi: 10.3321/j.issn:1001-7488.2009.07.028 |
|
| 谢瑾燕, 周健, 黄大庄. 基于RNA-seq数据开发梨网蝽SSR引物研究. 林业与生态科学, 2019, 34 (3): 280- 285. | |
| Xie J Y , Zhou J , Huang D Z . Development of SSR primers for Stephanitis nashi based on its RNA-seq datasets. Forestry and Ecological Science, 2019, 34 (3): 280- 285. | |
| 熊翠玲, 张璐, 付中民, 等. 基于RNA-seq数据大规模开发中华蜜蜂幼虫的SSR分子标记. 环境昆虫学报, 2017, 39 (1): 68- 74. | |
| Xiong C L , Zhang L , Fu Z M , et al. Large-scale development of SSR primers for Apis cerana cerana larvae based on its RNA-seq datasets. Journal of Environmental Entomology, 2017, 39 (1): 68- 74. | |
| 杨纤纤. 2017. 桉树枝瘿姬小蜂生殖生物学研究. 广州: 华南农业大学硕士学位论文. | |
| Yang X X. 2017. Reproductive biology of Leptocybe invasa Fisher & LaSalle. Guangzhou: MS thesis of South China Agricultural University. [in Chinese] | |
| 曾凡玉, 袁长宏, 陈元生. 赣南桉树枝瘿姬小蜂的发生与防治. 森林保护, 2015, (7): 51- 54. | |
| Zeng F Y , Yuan C H , Chen Y S . Occurrence and control of Leptocybe invasa in southern Ganzhou. Forest Protection, 2015, (7): 51- 54. | |
| 张华峰. 2013. 桉树枝瘿姬小蜂侵害机理及寄主桉树化学防御研究. 福州: 福建农林大学博士学位论文. | |
| Zhang H F. 2013. Studies on mechanism of Leptocybe invasa infestation and chemical defense of Eucalyptus. Fuzhou: PhD thesis of Fujian Agriculture and Forestry University. [in Chinese] | |
| 张华峰, 陈顺立, 康文通, 等. 桉树枝瘿姬小蜂对不同桉树品系及颜色的选择. 浙江农林大学学报, 2013, 30 (6): 904- 909. | |
| Zhang H F , Chen S L , Kang W T , et al. Selection behavior of Leptocybe invasa with different Eucalyptus cultivars and colors. Journal of Zhejiang Agriculture and Forestry University, 2013, 30 (6): 904- 909. | |
| 张鹏飞, 周晓榕, 庞保平, 等. 基于转录组数据高通量发掘沙葱萤叶甲微卫星引物. 应用昆虫学报, 2016, 53 (5): 1058- 1064. | |
| Zhang P F , Zhou X R , Pang B P , et al. High-throughput discovery of microsatellite markers in Galeruca daurica (Coleoptera: Chrysomelidae) from a transcriptome database. Chinese Journal of Applied Entomology, 2016, 53 (5): 1058- 1064. | |
| 朱方丽, 邱宝利, 任顺祥. 桉树枝瘿姬小蜂连续世代种群生命表. 生态学报, 2013, 33 (1): 97- 102. | |
| Zhu F L , Qiu B L , Ren S X . The continuous life-table of Leptocybe invasa. Acta Ecologica Sinica, 2013, 33 (1): 97- 102. | |
| Bérubé Y , Zhuang J , Rungis D , et al. Characterization of EST-SSRs in loblolly pine and spruce. Tree Genetics & Genomes, 2007, 3 (3): 251- 259. | |
|
Dittrich-Schröder G , Hoareau T B , Hurley B P , et al. Population genetic analyses of complex global insect invasions in managed landscapes: a Leptocybe invasa(Hymenoptera) case study. Biological Invasions, 2018, 20 (9): 2395- 2420.
doi: 10.1007/s10530-018-1709-0 |
|
|
Dittrich-Schröder G , Hurley B P , Wingfield M J , et al. Invasive gall-forming wasps that threaten non-native plantation-grown Eucalyptus: diversity and invasion patterns. Agricultural and Forest Entomology, 2020, 22 (4): 285- 297.
doi: 10.1111/afe.12402 |
|
|
Doğanlar O . Occurrence of Leptocybe invasa Fisher & La Salle, 2004(Hymenoptera: Chalcidoidea: Eulophidae) on Eucalyptus camaldulensis in Turkey, with description of the male sex. Zoology in the Middle East, 2005, 35 (1): 112- 114.
doi: 10.1080/09397140.2005.10638116 |
|
| Folmer O , Black M , Hoeh W , et al. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 1994, 3 (5): 294- 299. | |
|
Gupta A , Poorani J . Taxonomic studies on a collection of Chalcidoidea(Hymenoptera) from India with new distribution records. Journal of Threatened Taxa, 2009, 1 (5): 300- 304.
doi: 10.11609/JoTT.o1861.300-4 |
|
| Gupta P K , Balyan H S , Sharma P C , et al. Microsatellites in plants: a new class of molecular markers. Current Science, 1996, 70 (1): 45- 54. | |
|
Hu J , Zhang X Y , Jiang Z L , et al. New putative cryptic species detection and genetic network analysis of Bemisia tabaci(Hempitera: Aleyrodidae) in China based on mitochondrial COI sequences. Mitochondrial DNA Part A, 2018, 29 (3): 474- 484.
doi: 10.1080/24701394.2017.1307974 |
|
|
Hurley B P , Garnas J , Wingfield M J , et al. Increasing numbers and intercontinental spread of invasive insects on eucalypts. Biological Invasions, 2016, 18 (4): 921- 933.
doi: 10.1007/s10530-016-1081-x |
|
|
Kumar S , Stecher G , Tamura K . MEGA7: molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Molecular Biology and Evolution, 2016, 33 (7): 1870- 1874.
doi: 10.1093/molbev/msw054 |
|
|
Le N H , Nahrung H F , Griffiths M , et al. Invasive Leptocybe spp. and their natural enemies: global movement of an insect fauna on eucalypts. Biological Control, 2018, 125, 7- 14.
doi: 10.1016/j.biocontrol.2018.06.004 |
|
|
Lin C P , Danforth B N . How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined data sets. Molecular Phylogenetics and Evolution, 2004, 30 (3): 686- 702.
doi: 10.1016/S1055-7903(03)00241-0 |
|
| Madesis P , Ganopoulos I , Tsaftaris A . Microsatellites: evolution and contribution. Methods in Molecular Biology, 2013, 1006, 1- 13. | |
|
Mapondera T S , Burgess T , Matsuki M , et al. Identification and molecular phylogenetics of the cryptic species of the Gonipterus scutellatus complex(Coleoptera: Curculionidae: Gonipterini). Australian Journal of Entomology, 2012, 51 (3): 175- 188.
doi: 10.1111/j.1440-6055.2011.00853.x |
|
|
Mendel Z , Protasov A , Fisher N , et al. Taxonomy and biology of Leptocybe invasa gen. & sp. n. (Hymenoptera: Eulophidae), an invasive gall inducer on Eucalyptus. Australian Journal of Entomology, 2004, 43 (2): 101- 113.
doi: 10.1111/j.1440-6055.2003.00393.x |
|
|
Nugnes F , Gebiola M , Monti M M , et al. Genetic diversity of the invasive Gall Wasp Leptocybe invasa(Hymenoptera: Eulophidae) and of its Rickettsia endosymbiont, and associated sex-ratio differences. PLoS ONE, 2015, 10 (5): e0124660.
doi: 10.1371/journal.pone.0124660 |
|
|
Saha M C , Mian M A R , Eujayl I , et al. Tall fescue EST-SSR markers with transferability across several grass species. Theoretical and Applied Genetics, 2004, 109 (4): 783- 791.
doi: 10.1007/s00122-004-1681-1 |
|
|
Schröder M L , Slippers B , Wingfield M J , et al. Invasion history and management of Eucalyptus snout beetles in the Gonipterus scutellatus species complex. Journal of Pest Science, 2020, 93 (1): 11- 25.
doi: 10.1007/s10340-019-01156-y |
|
|
Scott K D , Eggler P , Seaton G , et al. Analysis of SSRs derived from grape ESTs. Theoretical and Applied Genetics, 2000, 100 (5): 723- 726.
doi: 10.1007/s001220051344 |
|
|
Tautz D , Renz M . Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Research, 1984, 12 (10): 4127- 4138.
doi: 10.1093/nar/12.10.4127 |
|
|
Temnykh S , Declerck G , Lukashova A , et al. Computational and experimental analysis of microsatellites in rice(Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Research, 2001, 11 (8): 1441- 1452.
doi: 10.1101/gr.184001 |
|
|
Untergasser A , Cutcutache I , Koressaar T , et al. Primer 3—new capabilities and interfaces. Nucleic Acids Research, 2012, 40 (15): e115.
doi: 10.1093/nar/gks596 |
|
|
Zhang D , Gao F L , Jakovli Ac'1 I , et al. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 2020, 20 (1): 348- 355.
doi: 10.1111/1755-0998.13096 |
|
|
Zheng X L , Li J , Yang Z D , et al. A review of invasive biology, prevalence and management of Leptocybe invasa Fisher & La Salle(Hymenoptera: Eulophidae: Tetrastichinae). African Entomology, 2014, 22 (1): 68- 79.
doi: 10.4001/003.022.0133 |
|
|
Zheng X L , Huang Z Y , Li J , et al. Reproductive biology of Leptocybe invasa Fisher & La Salle(Hymenoptera: Eulophidae). Neotropical Entomology, 2018a, 47 (1): 19- 25.
doi: 10.1007/s13744-017-0502-6 |
|
|
Zheng X L , Lin K , Huang Z Y , et al. Offspring sex ratio and reproductive tactics of Leptocybe invasa(Hymenoptera: Eulophidae): testing the effect of environmental characteristics. International Journal of Tropical Insect Science, 2018b, 38 (4): 394- 399.
doi: 10.1017/S1742758418000127 |
| [1] | 蔡金峰,杨晓明,郁万文,汪贵斌,曹福亮. 基于苦楝转录组测序的SSR分子标记开发[J]. 林业科学, 2021, 57(6): 85-92. |
| [2] | 毛伟兵, 陈发菊, 王长兰, 梁宏伟. 楸树雄性不育花芽转录组测序及分析[J]. 林业科学, 2017, 53(6): 141-150. |
| [3] | 唐海霞, 杜淑辉, 邢世岩, 桑亚林, 李际红, 刘晓静, 孙立民. 银杏性别决定相关基因的筛选[J]. 林业科学, 2017, 53(2): 76-82. |
| [4] | 林开勤, 赵德刚, 李岩, 何选泽, 王韶敏. 杜仲性别相关EST-SSR标记的开发[J]. 林业科学, 2016, 52(10): 146-152. |
| [5] | 张振, 张含国, 莫迟, 张磊. 红松转录组SSR分析及EST-SSR标记开发[J]. 林业科学, 2015, 51(8): 114-120. |
| [6] | 陈昊, 谭晓风. 基于油脂合成期油桐种仁转录组数据的α-亚麻酸代谢途径解析[J]. 林业科学, 2015, 51(3): 41-48. |
| [7] | 文亚峰, 韩文军, 周宏, 徐刚标. 杉木转录组SSR挖掘及EST-SSR标记规模化开发[J]. 林业科学, 2015, 51(11): 40-49. |
| [8] | 孙颖, 谭晓风, 罗敏, 李建安. 油桐花芽2个不同发育时期转录组分析[J]. 林业科学, 2014, 50(5): 70-74. |
| [9] | 钮世辉, 袁虎威, 陈晓阳, 李伟. 油松雌雄球花高通量基因表达谱芯片分析[J]. 林业科学, 2013, 49(9): 46-51. |
| [10] | 周炎花;乔小燕;马春雷;乔婷婷;金基强;姚明哲;陈亮. 广西茶树地方品种遗传多样性和遗传结构的EST-SSR分析[J]. 林业科学, 2011, 47(3): 59-67. |
| [11] | 杨秀艳;孙晓梅;张守攻;谢允慧;韩华. 日本落叶松EST-SSR标记开发及二代优树遗传多样性分析[J]. 林业科学, 2011, 47(11): 52-58. |
| [12] | 吴耀军 蒋学建 李德伟 罗基同 周国福 常明山 杨忠岐. 我国发现1种重要的林业外来入侵害虫——桉树枝瘿姬小蜂(膜翅目:姬小蜂科)[J]. 林业科学, 2009, 12(7): 161-163. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||