

林业科学 ›› 2025, Vol. 61 ›› Issue (12): 34-48.doi: 10.11707/j.1001-7488.LYKX20250230
• 前沿热点 • 上一篇
周龙1,4,张麦芳2,董强2,朱慧男1,3,*( ),刘金良1,4,*(
),刘金良1,4,*( )
)
收稿日期:2025-04-14
修回日期:2025-09-08
出版日期:2025-12-25
发布日期:2026-01-08
通讯作者:
朱慧男,刘金良
E-mail:geldwxh@163.com;liujinliang2016@nwafu.edu.cn
基金资助:
Long Zhou1,4,Maifang Zhang2,Qiang Dong2,Huinan Zhu1,3,*( ),Jinliang Liu1,4,*(
),Jinliang Liu1,4,*( )
)
Received:2025-04-14
Revised:2025-09-08
Online:2025-12-25
Published:2026-01-08
Contact:
Huinan Zhu,Jinliang Liu
E-mail:geldwxh@163.com;liujinliang2016@nwafu.edu.cn
摘要:
目的: 以陕西黄龙山林区油松-辽东栎混交林为研究对象,并以油松和辽东栎纯林为对照,分析不同林分类型下土壤微生物源碳(活体碳和残体碳)变化特征,探究油松-辽东栎混交林相较于纯林对土壤微生物源碳积累的影响,阐明驱动其变化的关键微生物功能机制,揭示调控该过程的关键环境因子。方法: 采用磷脂脂肪酸(PLFA)法测定微生物活体碳含量,氨基糖法测定微生物残体碳含量;运用微生物宏基因组测序技术,与Carbohydrate-Active enZYmes Database (CAZyme)和Kyoto Encyclopedia of Genes and Genomes (KEGG)数据库进行比对,获得并分析微生物碳分解功能基因特征;借助Mantel test分析,探究碳分解功能基因、土壤养分和微生物源碳的互作关系。基于数据分布特性,采用参数检验与非参数检验相结合的方法进行组间差异显著性分析和多重比较;数据统计分析与可视化使用R v4.2.1软件完成。结果: 1) 与油松和辽东栎纯林相比,油松-辽东栎混交可显著提高细菌(革兰氏阳性菌37.5%~54.9%)、真菌(子囊菌和担子菌35.7%~42.5%、丛枝菌根真菌23.7%~95.8%、接合菌57.8%~89.9%)和微生物(37.7%~55.1%)活体碳含量。2) 油松-辽东栎混交可显著提高土壤细菌残体碳含量(45.0%~56.7%)和微生物残体碳含量(20.2%~28.4%),并提升细菌残体碳(44.3%~59.9%)、真菌残体碳(15.3%~30.6%)和微生物残体碳(19.3%~34.7%)对有机碳库的贡献。3) 油松-辽东栎混交林下分解植物源碳(半纤维素、纤维素和木质素)和细菌源碳(肽聚糖)的CAZyme酶基因相对丰度介于油松纯林和辽东栎纯林之间,分解几丁质的CAZyme酶基因相对丰度高于纯林;KEGG碳分解功能基因表现出相似趋势。4) 细菌残体碳含量与放线菌活体碳含量(R = 0.65,P< 0.01)、厚壁菌活体碳含量(R = 0.60,P< 0.01)、革兰氏阴性菌活体碳含量(R = 0.67,P< 0.01)显著正相关;真菌残体碳含量与子囊菌和担子菌活体碳含量(R = 0.53,P< 0.05)、丛枝菌根真菌活体碳含量(R = 0.63,P< 0.05)、接合菌活体碳含量(R = 0.74,P< 0.01)显著正相关;微生物活体碳含量和残体碳含量均受土壤速效磷(AP)含量的显著影响;真菌活体碳含量∶细菌活体碳含量比值和真菌残体碳含量∶细菌残体碳含量比值均分别与分解植物源碳的CAZyme酶基因相对丰度和分解细菌源碳的CAZyme酶基因相对丰度显著正相关(P < 0.01)。结论: 油松-辽东栎混交能够显著提高细菌、真菌及微生物活体碳和残体碳含量,提升细菌残体碳、真菌残体碳和微生物残体碳含量对有机碳库的贡献。油松-辽东栎混交林中CAZyme酶基因相对丰度和碳降解功能基因绝对丰度介于油松纯林和辽东栎纯林之间,真菌活体碳含量∶细菌活体碳含量比值和真菌残体碳含量∶细菌残体碳含量比值均受CAZyme酶基因相对丰度和碳降解功能基因绝对丰度的影响。土壤中磷的有效性是影响微生物活体碳和残体碳积累的关键因素。
中图分类号:
周龙,张麦芳,董强,朱慧男,刘金良. 油松纯林、辽东栎纯林与油松-辽东栎混交林土壤微生物源碳变化特征[J]. 林业科学, 2025, 61(12): 34-48.
Long Zhou,Maifang Zhang,Qiang Dong,Huinan Zhu,Jinliang Liu. Characteristics of Soil Microbial-Derived Carbon Changes in Pinus tabuliformis Forest, Quercus wutaishanica Forest, and Their Mixed Forest[J]. Scientia Silvae Sinicae, 2025, 61(12): 34-48.
表1
各林分类型信息"
| 林分类型 Stand type | 优势乔木 Dominant tree species | 平均胸径 Mean DBH/ cm | 平均树高 Mean tree height/m | 海拔 Elevation/ m | 坡向 Aspect | 坡度 Slope/ (°) | 郁闭度 Canopy density | 灌木优势种Dominant shrub species | 草本优势种Dominant grass species |
| 油松纯林 Pinus tabuliformis forests | 油松 Pinus tabuliformis | 21.41±4.33 | 12.28±0.28 | 1 202 | 西南206° Southwest 206° | 23 | 0.60 | 野蔷薇Rosa multiflora、白刺花Sophora davidii、忍冬Lonicera japonica | 细秆薹草Carex capillaris、黄精Polygonatum sibiricum |
| 辽东栎纯林 Quercus wutaishanica forests | 辽东栎 Quercus wutaishanica | 21.41±0.34 | 11.41±0.75 | 1 603 | 北4° North 4° | 27 | 0.75 | 忍冬Lonicera japonica、红瑞木Lonicera japonica、荚蒾Lonicera japonica | 细秆薹草Carex capillaris、茜草 Rubia cordifolia |
| 油松-辽东栎 混交林 Pinus tabuliformis- Quercus wutaishanica mixed forests | 油松 Pinus tabuliformis | 25.90±2.70 | 14.59±0.83 | 1 535 | 西258° West 258° | 15 | 0.75 | 灰栒子Lonicera japonica、忍冬Lonicera japonica、红瑞木Lonicera japonica | 细秆薹草Carex capillaris、白茅草Imperata cylindrica |
| 辽东栎 Quercus wutaishanica | 18.90±3.67 | 11.55±1.55 |
表2
不同林分类型土壤养分状况及化学计量比①"
| 土壤养分特征 Soil nutrient characteristics | 油松纯林 Pinus tabuliformis forests | 辽东栎纯林 Quercus wutaishanica forests | 油松-辽东栎混交林 P. tabuliformis-Q. wutaishanica mixed forests |
| SOC/ (g·kg?1) | 13.17±0.89a | 16.05±1.67 a | 13.95±0.50 a |
| TN/ (g·kg?1) | 1.00±0.04 a | 1.40±0.10 a | 1.47±0.23 a |
| TP/ (g·kg?1) | 0.53±0.05 a | 0.64±0.02 a | 0.69±0.03 a |
| AP/ (mg·kg?1) | 0.18±0.01b | 0.28±0.03a | 0.22±0.05a |
| NH4+-N/ (mg·kg?1) | 2.32±0.22b | 4.60±0.34a | 5.03±0.18a |
| NO3?-N/ (mg·kg?1) | 0.30±0.05ab | 0.25±0.03b | 0.42±0.05a |
| C∶N | 13.13±0.72a | 11.36±0.60b | 10.27±0.30ab |
| C∶P | 26.12±4.45a | 25.21±2.33a | 20.49±1.37a |
| N∶P | 0.45±0.04a | 0.31±0.02b | 0.29±0.04b |
表3
不同林分类型土壤微生物活体碳含量①"
| 微生物活体碳类别 Microbial PLFA biomass | 油松纯林 P. tabuliformisforests | 辽东栎纯林 Q. wutaishanica forests | 油松-辽东栎混交林 P. tabuliformis-Q. wutaishanica mixed forests |
| 放线菌门Actinobacteria/ (mg·kg?1) | 1.34±0.04b | 1.32±0.16b | 1.98±0.30a |
| 厚壁菌门Firmicutes/ (mg·kg?1) | 1.86±0.02b | 2.20±0.29b | 2.95±0.09a |
| 丛枝菌根真菌Arbuscular mycorrhizal fungi/ (mg·kg?1) | 0.24±0.01b | 0.38±0.06ab | 0.47±0.07a |
| 子囊菌和担子菌门Ascomycota and Basidiomycota/ (mg·kg?1) | 0.40±0.06a | 0.42±0.08a | 0.57±0.02a |
| 接合菌门Zygomycota/ (mg·kg?1) | 0.69±0.01b | 0.83±0.12b | 1.31±0.10a |
| 革兰氏阴性菌Gram-negative bacteria/ (mg·kg?1) | 3.44±0.33a | 3.65±0.51a | 5.38±0.85a |
| 革兰氏阳性菌Gram-positive bacteria/ (mg·kg?1) | 3.17±0.12b | 3.57±0.45ab | 4.91±0.70a |
| 细菌Bacteria/ (mg·kg?1) | 6.61±0.42b | 7.18±0.96ab | 10.29±1.54a |
| 真菌Fungi/ (mg·kg?1) | 1.48±0.08b | 1.74±0.23b | 2.58±0.12a |
| 真菌∶细菌Ratio of bacteria to fungi | 0.22±0.01a | 0.26±0.02a | 0.24±0.02a |
| 微生物Microbial/ (mg·kg?1) | 9.73±0.58b | 10.96±1.59ab | 15.09±2.06a |

图1
不同林分类型单位质量有机碳中微生物活体碳含量 A:革兰氏阴性菌磷脂脂肪酸Gram-negative bacterial PLFA;B:革兰氏阳性菌磷脂脂肪酸Gram-positive bacterial PLFA;C:细菌磷脂脂肪酸 Bacterial PLFA;D:真菌磷脂脂肪酸 Fungal PLFA;E:微生物磷脂脂肪酸 Microbial PLFA. 不同林分类型间不同小写字母表示差异显著(P<0.05)。Different lowercase letters between different stand types indicate significant differences (P<0.05)."
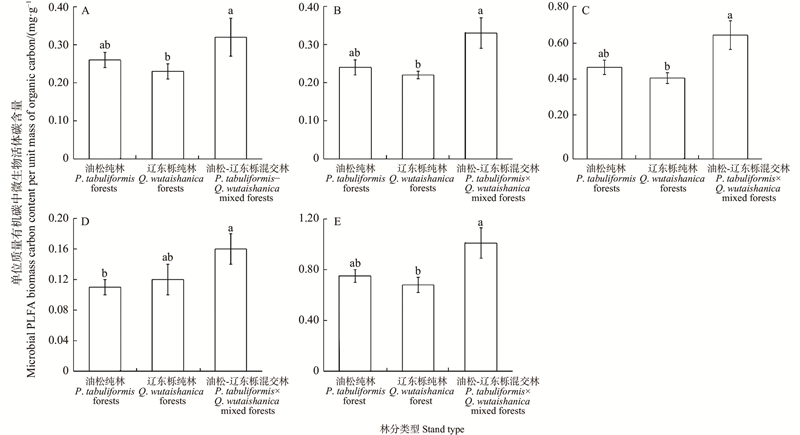
表4
不同林分类型土壤微生物残体碳及氨基糖含量①"
| 项目Item | 油松纯林 P. tabuliformis forests | 辽东栎纯林 Q. wutaishanica forests | 油松-辽东栎混交林 P. tabuliformis-Q. wutaishanica mixed forests |
| MurN/ (mg·kg?1) | 19.66±1.04b | 21.25±1.40b | 30.81±4.36a |
| GalN/ (mg·kg?1) | 209.88±13.00a | 298.43±20.27a | 302.59±47.23a |
| GluN/ (mg·kg?1) | 566.37±7.83b | 603.85±52.32ab | 709.70±10.67a |
| BNC/ (mg·kg?1) | 884.66±46.59b | 956.28±62.78b | 1 386.65±196.15a |
| FNC/ (mg·kg?1) | 4 844.90±57.31b | 5 161.81±455.46ab | 6 000.98±80.10a |
| MNC/ (mg·kg?1) | 5 729.49±103.60b | 6 118.09±510.16b | 7 354.68±136.04a |
| F∶BN | 5.53±0.24a | 5.39±0.26a | 4.69±0.68a |
表5
不同林分类型植物源和微生物源成分编码差异CAZyme基因相对丰度①"
| 组分Components | 油松纯林 P. tabuliformis forests | 辽东栎纯林 Q. wutaishanica forests | 油松-辽东栎混交林 P. tabuliformis-Q. wutaishanica mixed forests | |
| 植物源 Plant-derived | 半纤维素Hemicellulose | 48.49±0.43b | 51.93±1.07a | 49.86±0.73ab |
| 纤维素Cellulose | 7.40±0.16b | 8.09±0.09a | 8.01±0.17a | |
| 木质素Lignin | 30.56±0.32a | 27.75±0.86b | 29.44±0.49ab | |
| 总和Total | 86.45±0.09b | 87.76±0.30a | 87.31±0.27ab | |
| 细菌源 Bacteria-derived | 肽聚糖Peptidoglycan | 9.54±0.09a | 8.28±0.39b | 8.59±0.44ab |
| 真菌源 Fungi-derived | 几丁质Chitin | 2.93±0.11a | 2.79±0.05a | 3.02±0.16a |
| 葡聚糖Glucose | 1.08±0.06a | 1.16±0.13a | 1.08±0.07a | |
| 总和Total | 4.01±0.08a | 3.96±0.16a | 4.09±0.21a | |

图3
降解植物及微生物源碳的微生物CAZyme基因相对丰度聚类分析 CBM8:混合连接碳水化合物结合模块Hybrid connection carbohydrate binding module;GH1: β-葡萄糖苷酶 β-glucosidase;CBM63:纤维素碳水化合物结合模块 Cellulose carbohydrate binding module;GH3:纤维二糖β-葡萄糖苷酶 Cellobiose β-glucosidase;GH6:纤维二糖水解酶/内切葡聚糖酶 Cellobiohydrolase/endoglucanase;GH115:α-可溶性寡糖葡萄糖醛酸苷酶 α-soluble oligosaccharide glucuronidase;GH8:内切-β-1, 4-葡聚糖酶/木聚糖酶 Endo-β-1, 4-glucanase/xylanase;GH9:纤维二糖水解酶/内切葡聚糖酶 Cellobiohydrolase/endoglucanase;GH36:β-木糖苷酶 β- xylosidase;CE5:角质酶/乙酰木聚糖酯酶 Cutinase/acetyl xylan esterase;GH11:外切-β-1, 4-木聚糖酶 Exo-β-1, 4-xylanase;GH52:β-多糖-木糖苷酶 β-polysaccharide-xylosidase;GH26:β-甘露聚糖酶 β-mannanase;GH39:β-木聚糖-木糖苷酶 β-xylan-xylosidase;CE6:乙酰木聚糖酯酶 Acetyl xylan esterase;CE4:几丁质脱乙酰酶 Chitin deacetylase;GH43:β-木糖苷酶/α-阿拉伯呋喃糖苷酶 β-xylosidase/alpha-arabinofuranosidase;GH16:内切-β-1, 3(4)-葡聚糖酶/木葡聚糖酶 Endo-β-1, 3(4)-glucanase/xyloglucanase;GH120:β-木糖苷酶 β-xylosidase;GH62:α-专一阿拉伯呋喃糖苷酶 α-proprietary arabinofuranosidase;CE1:羧酸酯酶 Carboxylesterase;GH2:β-半乳糖苷酶 β-galactosidase;GH74:木葡聚糖特异性内切葡聚糖酶 Xyloglucan-specific endoglucanase;GH67:α-葡萄糖醛酸苷酶 α-glucuronidase;AA6:对苯醌还原酶 1,4-benzoquinone reductase;AA1:漆酶 Laccase;AA5:铜自由基氧化酶 Copper radical oxidase;AA3:葡萄糖-甲醇-胆碱氧化还原酶 Glucose-methanol-choline oxidoreductase;AA2:锰过氧化物酶 Manganese peroxidase;GH108:特异性溶菌酶 Specific lysozyme;GH25:桶状溶菌酶 Barrel lysozyme;GH104:λ-溶菌酶 λ-lysozyme;GH23:经典溶菌酶 Classic lysozyme;GH102:切肽链溶菌酶 Cleaved peptide chain lysozyme;CBM5:外切几丁质碳水化合物结合模块 Exfoliated chitin carbohydrate binding module;GH18:外切几丁质酶 Exonuclease;GH20:β- N-乙酰己糖胺酶 β- N-acetylhexosaminidase;GH81:β-1, 3-内切葡聚糖酶 β-1, 3-endoglucanase;GH128:线性β-1, 3-葡聚糖酶 Linear β-1, 3-glucanase;GH64:线性β-1, 3-葡聚糖 Linear β-1, 3-glucan;GH17:β-1, 3-葡聚糖酶 β-1, 3-glucanase. *:P<0.05;**:P<0.01;***: P<0.001."
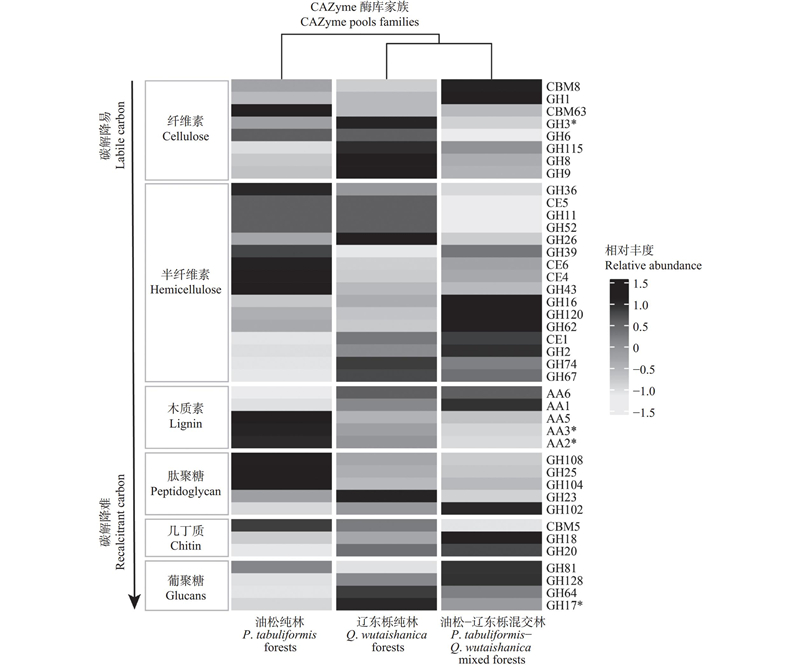
表6
不同林分类型植物源和微生物源成分编码差异碳降解功能基因绝对丰度①"
| 组分Components | 油松纯林 P. tabuliformis forests | 辽东栎纯林 Q. wutaishanica forests | 松栎油松-辽东栎混交林 P. tabuliformis-Q. wutaishanica mixed forests | |
| 淀粉 Starch | AMY | 2.93±0.01a | 2.92±0.01ab | 2.90±0.01b |
| GBE | 2.85±0.01b | 2.90±0.01a | 2.89±0.01a | |
| GP | 3.02±0.01b | 3.11±0.02a | 3.03±0.01b | |
| 4-α-GT | 2.73±0.01ab | 2.81±0.01a | 2.74±0.01b | |
| OLG | 1.91±0.03b | 2.09±0.05a | 1.97±0.03ab | |
| 纤维素 Cellulose | β-GLU | 2.75±0.02a | 2.91±0.02ab | 2.79±0.03b |
| α-D-XYL | 2.05±0.02b | 2.14±0.02a | 2.06±0.03b | |
| 半纤维素 Hemicellulose | EX | 2.02±0.04a | 1.96±0.08ab | 1.83±0.04b |
| β-MAN | 1.88±0.02c | 2.10±0.03b | 1.99±0.02a | |
| α-GAL | 2.28±0.01b | 2.47±0.03a | 2.39±0.03a | |
| β-GAL | 1.95±0.02a | 2.11±0.06ab | 2.12±0.06b | |
| α-L-FUC2 | 2.02±0.01a | 2.03±0.04a | 1.84±0.05b | |
| 木质素 Lignin | TYR | 2.14±0.02b | 2.10±0.03b | 2.04±0.03ab |
| CAT | 2.67±0.02b | 2.82±0.04a | 2.71±0.02b | |
| CPO | 2.54±0.01c | 2.71±0.03b | 2.61±0.03a | |
| NQR | 3.14±0.01b | 3.19±0.01a | 3.14±0.01b | |
| 果胶 Pectin | α-L-RHA | 2.23±0.02a | 2.11±0.06b | 2.25±0.02a |
| 芳香族化合物 Aromatic | MIL | 1.93±0.04b | 2.05±0.02a | 2.00±0.04ab |

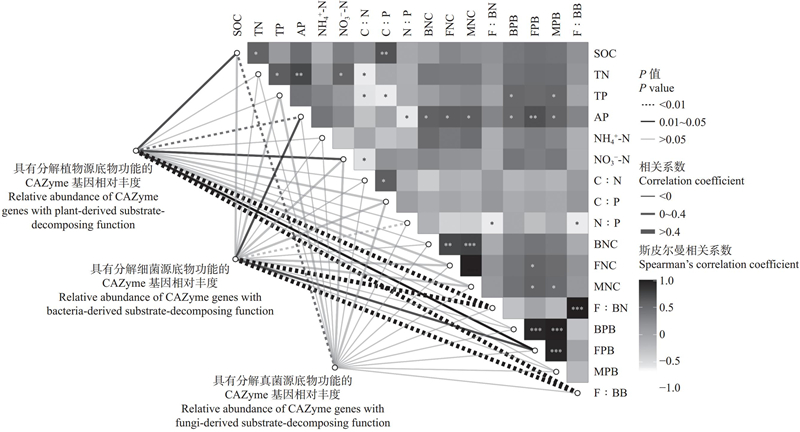
图4
分解植物源、细菌源及真菌源底物的CAZymes基因相对丰度与微生物量碳及环境因子的关联性分析 SOC:有机碳含量 Soil organic carbon content;TN:全氮含量 Total nitrogen content;TP:全磷含量 Total phosphorus content;AP:速效磷含量 Available phosphorus content;NH4+-N:铵态氮含量 Ammonium nitrogen content;NO3?-N:硝态氮含量 Nitrate nitrogen content;C∶N:有机碳含量∶全氮含量 Ratio of soil organic carbon content to total nitrogen content;C∶P:有机碳含量∶全磷含量Ratio of soil organic carbon content to total phosphorus content;N∶P:全氮含量∶全磷含量Ratio of total nitrogen content to total phosphorus content;BNC:细菌残体碳含量 Bacterial necromass carbon content;FNC:真菌残体碳含量 Fungal necromass carbon content;MNC:微生物残体碳含量 Microbial necromass carbon content;F∶BN:真菌残体碳含量∶细菌残体碳含量比值 Ratio of fungal necromass carbon to bacterial necromass carbon;BPB:细菌活体碳含量 Bacterial PLFA biomass carbon content;FPB:真菌活体碳含量 Fungal PLFA biomass carbon content;MPB:微生物活体碳含量 Microbial PLFA biomass carbon content;F:BB:真菌活体碳含量:细菌活体碳含量比值Ratio of fungal PLFA biomass carbon to bacterial PLFA biomass carbon. *:P<0.05;**:P<0.01;***: P<0.001."
|
白昱欣, 刘润洪, 苏洁桦, 等. 树种混交对马尾松和红锥根际与非根际土壤微生物资源限制的影响. 生态学报, 2024, 44 (23): 10770- 10781.
doi: 10.20103/j.stxb.202310312359 |
|
|
Bai Y X, Liu R H, Su J H, et al. Effects of tree species mixing on bulk and rhizosphere soil microbial resource limitation in stands of Pinus massoniana and Castanopsis hystrix. Acta Ecologica Sinica, 2024, 44 (23): 10770- 10781.
doi: 10.20103/j.stxb.202310312359 |
|
| 淡彩虹. 2023. 黄土高原人工林混交模式对土壤团聚体稳定性及有机碳的影响. 杨凌: 西北农林科技大学. | |
| Dan C H. 2023. Effects of artificial mixed forest models on soil aggregate stability and organic carbon in the Loess Plateau. Yangling: Northwest A&F University. [in Chinese] | |
| 巩 晨. 2022. 黄土高原不同造林模式生态功能特征及权衡/协同效应. 杨凌: 中国科学院大学(中国科学院教育部水土保持与生态环境研究中心). | |
| Gong C. 2022. Characteristics and trade-off/synergy effects of ecological functions under different afforestation patterns on China’s Loess Plateau. Yangling: University of Chinese Academy of Sciences (Institute of Soil and Water Conservation Chinese Academy of Sciences and Ministry of Water Resources). [in Chinese] | |
|
樊容源, 叶绍明, 张钱春, 等. 杉木纯林及其混交林土壤团聚体活性有机碳组分特征. 西北林学院学报, 2023, 38 (6): 20- 28, 37.
doi: 10.3969/j.issn.1001-7461.2023.06.03 |
|
|
Fan R Y, Ye S M, Zhang Q C, et al. Characteristics of active organic carbon components in soil aggregates of pure plantation of C.lanceolata and its mixed forest. Journal of Northwest Forestry University, 2023, 38 (6): 20- 28, 37.
doi: 10.3969/j.issn.1001-7461.2023.06.03 |
|
| 芦 月, 杨 静, 李仕辉, 等. 2025. 黄河三角洲不同经济林土壤有机碳分布特征及改良土壤效应. 中国水土保持科学(中英文), 23(2): 180−189. | |
| Lu Y, Yang J, Li S H, et al. 2025. Distribution characteristics of soil organic carbon and soil improvement effects of different economic forests in the Yellow River Delta. Science of Soil and Water Conservation, 23(2): 180−189. [in Chinese] | |
|
王小平, 杨 雪, 杨 楠, 等. 凋落物多样性及组成对凋落物分解及土壤微生物群落的影响: 二氧化碳倍增条件下. 生态学报, 2020, 40 (17): 6171- 6178.
doi: 10.5846/stxb201812162743 |
|
|
Wang X P, Yang X, Yang N, et al. Effects of litter diversity and composition on litter decomposition characteristics and soil microbial community: under the conditions of doubling ambient atmospheric CO2 concentration. Acta Ecologica Sinica, 2020, 40 (17): 6171- 6178.
doi: 10.5846/stxb201812162743 |
|
|
严明明, 高洪娣, 刘美华, 等. 混交改造杉木纯林对林下土壤肥力影响的研究进展. 生态科学, 2024, 43 (2): 253- 259.
doi: 10.14108/j.cnki.1008-8873.2024.02.028 |
|
|
Yan M M, Gao H D, Liu M H, et al. Research progress on the effect of mixed plantation on the soil fertility in theforests between Cunninghamia lanceolata and other tree species. Ecological Science, 2024, 43 (2): 253- 259.
doi: 10.14108/j.cnki.1008-8873.2024.02.028 |
|
|
杨洪炳, 肖文发, 曾立雄, 等. 2种马尾松混交林土壤微生物特性及其与团聚体稳定性耦合关系. 江西农业大学学报, 2024, 46 (3): 543- 557.
doi: 10.3724/aauj.2024049 |
|
|
Yang H B, Xiao W F, Zeng L X, et al. Microbial characteristics in soil aggregates and their coupling with aggregates stability in two species of mixed Pinus massoniana forests. Acta Agriculturae Universitatis Jiangxiensis, 2024, 46 (3): 543- 557.
doi: 10.3724/aauj.2024049 |
|
|
姚小萌, 牛桠枫, 党珍珍, 等. 黄土高原自然植被恢复对土壤质量的影响. 地球环境学报, 2015, 6 (4): 238- 247.
doi: 10.7515/JEE201504006 |
|
|
Yao X M, Niu Y F, Dang Z Z, et al. Effects of natural vegetation restoration on soil quality on the Loess Plateau. Journal of Earth Environment, 2015, 6 (4): 238- 247.
doi: 10.7515/JEE201504006 |
|
| 于志静. 2024. 黄土丘陵区刺槐-侧柏混交林对土壤有机碳及其矿化的调控机制. 杨凌: 西北农林科技大学. | |
| Yu Z J. 2024. The regulatory mechanism of Robinia pseudoacacia-Platycladus orientalis mixed forests on soil organic carbon and its mineralization in the Loess Hilly Region. Yangling: Northwest A&F University. [in Chinese] | |
|
周建云, 李 荣, 何景峰, 等. 近自然经营对辽东栎林优势乔木更新的影响. 林业科学, 2013, 49 (8): 15- 20.
doi: 10.11707/j.1001-7488.20130803 |
|
|
Zhou J Y, Li R, He J F, et al. Regeneration of the Dominant Arbors after Close-to-Natural Management of Quercus wutaishanica Forest. Scientia Silvae Sinicae, 2013, 49 (8): 15- 20.
doi: 10.11707/j.1001-7488.20130803 |
|
|
张维伟, 尹代皓, 雷雨婷, 等. 黄土高原南部麻栎不同群落类型乔木种群结构及演替模拟. 生态学报, 2024, 44 (6): 2572- 2581.
doi: 10.20103/j.stxb.202110263011 |
|
|
Zhang W W, Yin D H, Lei Y T, et al. Structural dynamics and succession simulation of the dominant populations in three types of Quercus acutissima community in southern of the Losses Plateau. Acta Ecologica Sinica, 2024, 44 (6): 2572- 2581.
doi: 10.20103/j.stxb.202110263011 |
|
|
张兹箕, 王启鑫, 赵振宇, 等. 鲁中南地区不同经济林种植模式的改良土壤效应. 水土保持研究, 2024, 31 (5): 102- 111.
doi: 10.13869/j.cnki.rswc.2024.05.021 |
|
|
Zhang Z J, Wang Q X, Zhao Z Y, et al. Soil Improvement Effect of Different Economic Forest Planting Modes in South-central Shandong Province. Research of Soil and Water Conservation, 2024, 31 (5): 102- 111.
doi: 10.13869/j.cnki.rswc.2024.05.021 |
|
|
Azeem M, Sun T R, Jeyasundar P G S A, et al. Biochar-derived dissolved organic matter (BDOM) and its influence on soil microbial community composition, function, and activity: a review. Critical Reviews in Environmental Science and Technology, 2023, 53 (21): 1912- 1934.
doi: 10.1080/10643389.2023.2190333 |
|
|
Bourget M Y, Fanin N, Fromin N, et al. Plant litter chemistry drives long-lasting changes in the catabolic capacities of soil microbial communities. Functional Ecology, 2023, 37 (7): 2014- 2028.
doi: 10.1111/1365-2435.14379 |
|
|
Cudjoe E, Ruiz-Peinado R, Pretzsch H, et al. Neighborhood competition improves biomass estimation for Scots pine (Pinus sylvestris L.) but not Pyrenean oak (Quercus pyrenaica Willd. ) in young mixed forest stands. Forest Ecosystems, 2025, 13, 100317..
doi: 10.1016/j.fecs.2025.100317 |
|
|
Choreño-Parra E M, Treseder K K. Mycorrhizal fungi modify decomposition: a meta-analysis. New Phytologist, 2024, 242 (6): 2763- 2774.
doi: 10.1111/nph.19748 |
|
|
Duarte A G, Maherali H. A meta-analysis of the effects of climate change on the mutualism between plants and arbuscular mycorrhizal fungi. Ecology and Evolution, 2022, 12 (1): e8518.
doi: 10.1002/ece3.8518 |
|
|
Hu D, Xu Y, Chai Y F, et al. Spatial Distribution pattern and genetic diversity of Quercus wutaishanica Mayr population in Loess Plateau of China. Forests, 2022, 13 (9): 1375.
doi: 10.3390/f13091375 |
|
|
Fan X Y, Ge A H, Qi S S, et al. Root exudates and microbial metabolites: signals and nutrients in plant-microbe interactions. Science China Life Sciences, 2025, 68, 2290- 2302.
doi: 10.1007/s11427-024-2876-0 |
|
|
Fu X H, Song Q L, Li S Q, et al. Dynamic changes in bacterial community structure are associated with distinct priming effect patterns. Soil Biology and Biochemistry, 2022, 169, 108671.
doi: 10.1016/j.soilbio.2022.108671 |
|
|
Gedminienė L, Spiridonov A, Stančikaitė M, et al. Temporal and spatial climate changes in the mid-Baltic region in the Late Glacial and the Holocene: Pollen-based reconstructions. Catena, 2025, 252, 108851.
doi: 10.1016/j.catena.2025.108851 |
|
|
Gao G, Huang X M, Xu H C, et al. Conversion of pure Chinese fir plantation to multi-layered mixed plantation enhances the soil aggregate stability by regulating microbial communities in subtropical China. Forest Ecosystems, 2022, 9, 100078.
doi: 10.1016/j.fecs.2022.100078 |
|
|
Han Z G, Zhou Y C, Guo Y L, et al. Organic carbon sequestration by Fe oxides in aggregates in a Pinus massoniana conifer-broadleaf mixed forest. Soil and Tillage Research, 2024, 240, 106105.
doi: 10.1016/j.still.2024.106105 |
|
|
Hu D, Zhou X H, Ma G Y, et al. Increased soil bacteria-fungus interactions promote soil nutrient availability, plant growth, and coexistence. Science of The Total Environment, 2024, 955, 176919.
doi: 10.1016/j.scitotenv.2024.176919 |
|
|
Huffman M S, Madritch M D. Soil microbial response following wildfires in thermic oak-pine forests. Biology and Fertility of Soils, 2018, 54, 985- 997.
doi: 10.1007/s00374-018-1322-5 |
|
|
Jiang F Y, Zhang L, Zhou J C, et al. Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae. New Phytologist, 2021, 230, 304- 315.
doi: 10.1111/nph.17081 |
|
|
Joergensen R G. Amino sugars as specific indices for fungal and bacterial residues in soil. Biology and Fertility of Soils, 2018, 54 (5): 559- 568.
doi: 10.1007/s00374-018-1288-3 |
|
|
Kakouridis A, Yuan M T, Nuccio E E, et al. Arbuscular mycorrhiza convey significant plant carbon to a diverse hyphosphere microbial food web and mineral-associated organic matter. New Phytologist, 2024, 242, 1661- 1675.
doi: 10.1111/nph.19560 |
|
|
Kohler J, Roldán A, Campoy M, et al. Unraveling the role of hyphal networks from arbuscular mycorrhizal fungi in aggregate stabilization of semiarid soils with different textures and carbonate contents. Plant and Soil, 2017, 410, 273- 281.
doi: 10.1007/s11104-016-3001-3 |
|
|
Liang C, Amelung W, Lehmann J, et al. Quantitative assessment of microbial necromass contribution to soil organic matter. Global Change Biology, 2019, 25 (11): 3578- 3590.
doi: 10.1111/gcb.14781 |
|
|
Mercer G D, Mickan B S, Gleeson D B, et al. Probing the pump: soil carbon dynamics, microbial carbon use efficiency and community composition in response to stoichiometrically-balanced compost and biochar. Soil Biology and Biochemistry, 2025, 205, 109770..
doi: 10.1016/j.soilbio.2025.109770 |
|
|
Manici L M, Caputo F, Fornasier F, et al. Ascomycota and Basidiomycota fungal phyla as indicators of land use efficiency for soil organic carbon accrual with woody plantations. Ecological Indicators, 2024, 160, 111796.
doi: 10.1016/j.ecolind.2024.111796 |
|
|
Mou Z J, Kuang L H, Zhang J, et al. Nutrient availability and stoichiometry mediate microbial effects on soil carbon sequestration in tropical forests. Soil Biology and Biochemistry, 2023, 186, 109186.
doi: 10.1016/j.soilbio.2023.109186 |
|
|
Starke R, Mondéjar R L, Human Z R, et al. Niche differentiation of bacteria and fungi in carbon and nitrogen cycling of different habitats in a temperate coniferous forest: a metaproteomic approach. Soil Biology and Biochemistry, 2021, 155, 108170.
doi: 10.1016/j.soilbio.2021.108170 |
|
|
Tao F, Huang Y Y, Hungate B A, et al. Microbial carbon use efficiency promotes global soil carbon storage. Nature, 2023, 618, 981- 985.
doi: 10.1038/s41586-023-06042-3 |
|
|
Toriyama J, Hashimoto S, Nakao K, et al. Management strategies for shrinking and aging tree plantations are constrained by the synergies and trade-offs between carbon sequestration and other forest ecosystem services. Journal of Environmental Management, 2025, 373, 123762.
doi: 10.1016/j.jenvman.2024.123762 |
|
|
Wang B R, Huang Y M, Li N, et al. Initial soil formation by biocrusts: Nitrogen demand and clay protection control microbial necromass accrual and recycling. Soil Biology and Biochemistry, 2022, 167, 108607.
doi: 10.1016/j.soilbio.2022.108607 |
|
|
Wiesenbauer J, Gorka S, Jenab K, et al. Preferential use of organic acids over sugars by soil microbes in simulated root exudation. Soil Biology and Biochemistry, 2025, 203, 109738.
doi: 10.1016/j.soilbio.2025.109738 |
|
|
Zhang L M, Wu Y N, Yan L B, et al. Modeling the maximal organic carbon saturation capacity in karst forest soils of China: comparing the existing models. Applied Ecology and Environmental Research, 2021, 19 (5): 4115- 4128.
doi: 10.15666/aeer/1905_41154128 |
| [1] | 孙英杰,张德楠,沈育伊,徐广平,曹杨,黄科朝,陈运霜,毛馨月,滕秋梅,吕仕洪,褚俊智. 模拟氮沉降对中亚热带桉树人工林土壤微生物群落结构及酶活性的影响[J]. 林业科学, 2025, 61(5): 46-60. |
| [2] | 李晓,贾淑娴,席颖青,杨柳明,刘小飞. 凋落物添加与去除对米槠天然林土壤微生物残体碳的影响[J]. 林业科学, 2024, 60(10): 12-20. |
| [3] | 崔朝伟,彭丽鸿,马东旭,王佳琪,江祥庆,江先桂,马祥庆,林开敏. 间伐对杉木人工林土壤微生物残体碳的影响[J]. 林业科学, 2023, 59(5): 41-52. |
| [4] | 贺同鑫, 孙建飞, 李艳鹏, 俞有志, 胡宝清, 王清奎. 环割对杉木和马尾松人工林土壤微生物群落结构的影响[J]. 林业科学, 2017, 53(6): 77-84. |
| [5] | 字洪标, 向泽宇, 王根绪, 阿的鲁骥, 王长庭. 青海不同林分土壤微生物群落结构(PLFA)[J]. 林业科学, 2017, 53(3): 21-32. |
| [6] | 刘彩霞, 焦如珍, 董玉红, 孙启武, 刘少文. 应用PLFA方法分析氮沉降对土壤微生物群落结构的影响[J]. 林业科学, 2015, 51(6): 155-162. |
| [7] | 张于光, 宿秀江, 丛静, 陈展, 卢慧, 刘敏超, 李迪强. 神农架土壤微生物群落的海拔梯度变化[J]. 林业科学, 2014, 50(9): 161-166. |
| [8] | 孙棣棣;徐秋芳;田甜;刘卜榕. 不同栽培历史毛竹林土壤微生物生物量及群落组成变化[J]. 林业科学, 2011, 47(7): 181-186. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||