

Scientia Silvae Sinicae ›› 2023, Vol. 59 ›› Issue (1): 110-118.doi: 10.11707/j.1001-7488.LYKX20210971
• Research papers • Previous Articles Next Articles
Shanshan Ma1,Jingjing Yang1,Dehui Qu1,Mengjie Li1,Jin Zhang1,Fanlin Wu1,Hongyan Su1,3,Lei Wang2,*
Received:2021-12-30
Online:2023-01-25
Published:2023-02-24
Contact:
Lei Wang
CLC Number:
Shanshan Ma,Jingjing Yang,Dehui Qu,Mengjie Li,Jin Zhang,Fanlin Wu,Hongyan Su,Lei Wang. Overexpression of PdbZFP26, a Gene Encoding C2H2 Zinc Finger Protein, Improves Salt Tolerance of Transgenic Populus davidiana×P. bolleana[J]. Scientia Silvae Sinicae, 2023, 59(1): 110-118.
Table 1
Primer sequence used in PCR"
| 用途 Application | 引物名称 Primer name | 引物序列 Sequence (5′—3′) |
| 载体构建 Vector construction | p2301-PdbZFP26-F | CGGGATCCATGCCAGTTGTGAAACCTGTGA |
| p2301-PdbZFP26-R | GCGTCGACTTAAATGATCGTTCTTTGTTTCGG | |
| 基因组扩增 Genome amplification | P35S | TCATTTGGAGAGAACACGGGG |
| Pzfp26-3 | CCAAAAGGTCAGAACCAGC | |
| 实时荧光定量PCR qRT-PCR | PActin-F | TCATCGGAATGGAAGCTGCTGGTA |
| PActin-R | TTAAATGATCGTTCTTTGTTTCGG | |
| Pzfp26-1 | GCCATTGCTGCTGAACATGTG | |
| Pzfp26-2 | AGCTACATTCGCAAGTCCAC | |
| Pzfp26-4 | GAGCCCGAGCTGATTACCTG | |
| Pzfp26-5 | CTAGTCCTCACCCTTCCCAT |
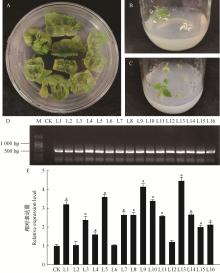
Fig.3
The screening and identification of transgenic P. davidiana×P. bolleana overexpressing PdbZFP26 gene A. Screening of resistant bud; B and C. Rooting screening; D. PCR detection of PdbZFP26 in genomic DNA of transgenic lines; E. Relative expression levels of PdbZFP26 in transgenic poplar lines."

|
黄骥, 王建飞, 张红生. 植物C2H2型锌指蛋白的结构与功能. 遗传, 2004, 26 (3): 414- 418.
doi: 10.3321/j.issn:0253-9772.2004.03.029 |
|
|
Huang J , Wang J F , Zhang H S . Structure and function of plant C2H2 zinc finger protein. Hereditas, 2004, 26 (3): 414- 418.
doi: 10.3321/j.issn:0253-9772.2004.03.029 |
|
| 李琳, 丁峰, 潘介春, 等. 植物锌指蛋白转录因子家族研究进展. 农业科学, 2020, 40 (2): 65- 75. | |
| Li L , Ding F , Pan J C , et al. Research progress on family of plant zinc-finger protein transcription factors. Chinese Journal of Tropical Agriculture, 2020, 40 (2): 65- 75. | |
|
刘福汉, 王遵亲. 潜水蒸发条件下不同质地剖面的土壤水盐运动. 土壤学报, 1993, 30 (2): 173- 181.
doi: 10.3321/j.issn:0564-3929.1993.02.003 |
|
|
Liu F H , Wang Z Q . Salt-water dynamics in soil profiles of different texture under ground water evaporation condition. Acta Pedologica Sinica, 1993, 30 (2): 173- 181.
doi: 10.3321/j.issn:0564-3929.1993.02.003 |
|
| 王利界, 周智彬, 常青, 等. 盐旱交叉胁迫对灰胡杨(Populus pruinosa)幼苗生长和生理生化特性的影响. 生态学报, 2018, 38 (19): 7026- 7033. | |
| Wang L J , Zhou Z B , Chang Q , et al. Effects of salt drought cross stress on growth and physiological and biochemical characteristics of Populus pruinosa seedlings. Acta Ecologica Sinica, 2018, 38 (19): 7026- 7033. | |
| 张佳, 刘俊芳, 赵婷婷, 等. 植物C2H2型锌指蛋白研究进展. 分子植物育种, 2018, 16 (2): 427- 433. | |
| Zhang J , Liu J F , Zhao T T , et al. Research progress of plant C2H2 zinc finger protein. Molecular Plant Breeding, 2018, 16 (2): 427- 433. | |
|
Agarwal P , Arora R , Ray S , et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Molecular Biology, 2007, 65 (4): 467- 485.
doi: 10.1007/s11103-007-9199-y |
|
|
Chappelle E W , Kim M S , McMurtrey J E . Ratio analysis of reflectance spectra (RARS): an algorithm for the remote estimation of the concentrations of chlorophyll-a, chlorophyll-b, and carotenoids in soybean leaves. Remote Sensing of Environment, 1992, 39, 239- 247.
doi: 10.1016/0034-4257(92)90089-3 |
|
|
Englbrecht C C , Schoof H , B hm S . Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics, 2004, 5 (1): 1- 17.
doi: 10.1186/1471-2164-5-1 |
|
| Gang L , Tai F J , Zheng Y , et al. Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Molecular Biology, 2010, 74 (4/5): 437- 452. | |
|
Hafeez M B , Zahra N , Zahra K , et al. Brassinosteroids: molecular and physiological responses in plant growth and abiotic stresses. Plant Stress, 2021, 2, e100029.
doi: 10.1016/j.stress.2021.100029 |
|
|
Han G , Yuan F , Guo J , et al. AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Science, 2019, 285, 55- 67.
doi: 10.1016/j.plantsci.2019.05.002 |
|
|
Huang J , Sun S , Xu D Q , et al. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Molecular Biology, 2012, 80 (3): 337- 350.
doi: 10.1007/s11103-012-9955-5 |
|
| Kanchan V , Neha U , Nitin K , et al. Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Frontiers in Plant Science, 2017, 8 (120): 161- 172. | |
|
Kodaira K S , Qin F , Tran L S P , et al. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiology, 2011, 157 (2): 742- 756.
doi: 10.1104/pp.111.182683 |
|
|
Li Y , Chu Z N , Luo J Y , et al. The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN5B. Plant Biotechnology Journal, 2018, 16 (6): 1201- 1203.
doi: 10.1111/pbi.12863 |
|
| Liu D C , Li Y , Luo M , et al. Molecular cloning and characterization of PtrZPT2-1, a ZPT2 family gene encoding a Cys2/His2-type zinc finger protein from trifoliate orange (Poncirus trifoliata (L.) Raf.) that enhances plant tolerance to multiple abiotic stresses. Plant Science, 2017, 49 (7): 66- 78. | |
|
Liu Q , Wang Z , Xu X , et al. Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa). PLoS One, 2015, 10 (8): e0134753.
doi: 10.1371/journal.pone.0134753 |
|
|
Livak K J , Schmittgen T D . Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 2001, 25 (4): 402- 408.
doi: 10.1006/meth.2001.1262 |
|
|
Lopato S , Bazanova N , Morran S , et al. Isolation of plant transcription factors using a modified yeast one-hybrid system. Plant Methods, 2006, 2 (1): 1- 15.
doi: 10.1186/1746-4811-2-1 |
|
|
Luo X , Cui N , Zhu Y M , et al. Over-expression of GsZFP1, an ABA-responsive C2H2-type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. Journal of Plant Physiology, 2012, 169 (12): 1192- 1202.
doi: 10.1016/j.jplph.2012.03.019 |
|
|
Ma X , Liang W , Gu P , et al. Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiology and Biochemistry, 2016, 106, 129- 140.
doi: 10.1016/j.plaphy.2016.04.033 |
|
|
Muthamilarasan M , Bonthala V S , Mishra A K , et al. C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Functional and Integrative Genomics, 2014, 14 (3): 531- 543.
doi: 10.1007/s10142-014-0383-2 |
|
|
Sun S J , Guo S Q , Xia Y , et al. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. Journal of Experimental Botany, 2010, 61 (10): 2807- 2818.
doi: 10.1093/jxb/erq120 |
|
|
Takatsuji H . Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Molecular Biology, 1999, 39 (6): 1073- 1078.
doi: 10.1023/A:1006184519697 |
|
|
Takatsuji H , Mori M , Benfey P N , et al. Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO Journal, 1992, 11 (1): 241- 249.
doi: 10.1002/j.1460-2075.1992.tb05047.x |
|
|
Wang F , Tong W , Zhu H , et al. A novel Cys2/His2 zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta, 2016, 243 (3): 783- 797.
doi: 10.1007/s00425-015-2443-9 |
|
|
Wolters H , Jurgens G . Survival of the flexible: hormonal growth control and adaptation in plant development. Nature Reviews Genetics, 2009, 10 (5): 305- 317.
doi: 10.1038/nrg2558 |
|
|
Xuan C N , Sun H , Hussain , et al. A positive transcription factor in osmotic stress tolerance, ZAT10, is regulated by MAP kinases in Arabidopsis. Journal of Plant Biology, 2016, 59 (1): 55- 61.
doi: 10.1007/s12374-016-0442-4 |
|
|
Xu D Q , Huang J , Guo S Q , et al. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Letters, 2008, 582 (7): 1037- 1043.
doi: 10.1016/j.febslet.2008.02.052 |
|
|
Yin J , Wang L , Zhao J , et al. Genome-wide characterization of the C2H2 zinc-finger genes in Cucumis sativus and functional analyses of four CsZFPs in response to stresses. BMC Plant Biology, 2020, 20 (1): 359- 380.
doi: 10.1186/s12870-020-02575-1 |
|
|
Yuan S , Li X , Li R , et al. Genome-wide identification and classification of Soybean C2H2 zinc finger proteins and their expression analysis in legume-rhizobium symbiosis. Frontiers in Microbiology, 2018, 9, 126- 144.
doi: 10.3389/fmicb.2018.00126 |
|
| Zhang Z Y , Liu H H , Sun C , et al. A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. Journal of Plant Physiology, 2018, 229, 100- 110. |
| [1] | Zhenqian Li,Xiaoqin Wu,Weiliang Kong. Effects of Interaction between Bacillus paramycoides JYZ-SD5 and Mycorrhizal Fungi on Growth and Salt Tolerance of Metasequoia glyptostroboides [J]. Scientia Silvae Sinicae, 2023, 59(1): 99-109. |
| [2] | Kai Zhao,Yan Fan,Shengqiang Zou,Xuhui Huan,Shuhui Du,Youzhi Han,Shengji Wang. Cloning and Expression Analysis of PagMYBR 96 Involved in Salt Stress Response in Populus alba × P. glandulosa '84K' [J]. Scientia Silvae Sinicae, 2022, 58(9): 117-127. |
| [3] | Qingzhi Lin,Xiangyuan Zhu,Peili Mao,Lin Zhu,Longmei Guo,Zexiu Li,Banghua Cao,Yingdong Hao,Haitao Tan,Pizheng Hong,Xiaojun Lu. Effects of NaCl and PEG Stresses on Germination and Seedling Growth of Robinia pseudoacacia Seeds with Different Sizes [J]. Scientia Silvae Sinicae, 2022, 58(2): 100-112. |
| [4] | Xiuting Zhao,Yanshuang Wang,Jie Duan,Lüyi Ma,Baohua He,Zhongkui Jia,Ziyang Sang,Zhonglong Zhu. Effects of Salt Stress on Growth and Photosynthetic Characteristics of Magnolia wufengensis Grafted Seedlings [J]. Scientia Silvae Sinicae, 2021, 57(4): 43-53. |
| [5] | Liu Daofeng, Wang Xia, Dai Yin, Yang Jianfeng, Ma Jing, Li Mingyang, Sui Shunzhao. Cloning and Function Analysis of CpTAF10 from Wintersweet (Chimonanthus praecox) [J]. Scientia Silvae Sinicae, 2019, 55(6): 176-183. |
| [6] | Jiang Cheng, Zhou Houjun, Zhao Yanqiu, He Hui, Chu Liwei, Song Xueqin, Lu Mengzhu. Effects of CDD Gene on the Growth and Development of Populus alba×P. glandulosa ‘84K’ in Response to Drought and Salt Stresses [J]. Scientia Silvae Sinicae, 2019, 55(2): 33-40. |
| [7] | Wang Kaili, Fu Ying, Zhou Mingbing. Identification and Expression Pattern of SQUAMOSA Promoter Binding Protein (SBP) Family Genes in Phyllostachys edulis [J]. Scientia Silvae Sinicae, 2017, 53(12): 50-61. |
| [8] | Chu Wenyuan, Wang Yujiao, Zhu Dongyue, Chen Zhu, Yan Hanwei, Xiang Yan. Selection of Novel Reference Genes in Poplar under Salt and Drought Stresses [J]. Scientia Silvae Sinicae, 2017, 53(10): 70-79. |
| [9] | Guan Yang, Wang Hongzhi, Zhang Jiewei, Chen Yajuan, Zhu Dan, Ding Liping, Wei Jianhua. Isolation and Expression of Eukaryotic Translation Initiation Factor 5A2 in Populus tomentosa [J]. Scientia Silvae Sinicae, 2014, 50(2): 63-69. |
| [10] | Yang Xiuyan, Zhang Huaxin, Zhang Li, Liu Zhengxiang, Yang Sheng, Wu Xiang. Effects of NaCl Stress on Growth and Absorption, Transportation and Distribution of Ions in Nitraria tangutorum [J]. Scientia Silvae Sinicae, 2013, 49(9): 165-171. |
| [11] | Ma Jinlong, Jiang Guobin, Yao Shanjing, Jin Hua. Effects of Apoplastic Ion Contents in the Tender Stems of Two Populus Cultivars on Gas Exchange Parameters Under Salt Stress [J]. Scientia Silvae Sinicae, 2013, 49(7): 40-47. |
| [12] | Yang Sheng;Liu Zhengxiang;Zhang Huaxin;Yang Xiuyan;Liu Tao;Yao Zongguo. Comprehensive Evaluation of Salt Tolerance and Screening Identification Indexes for Three Tree Species [J]. , 2013, 49(1): 91-98. |
| [13] | Wang Bin;Ju Bo;Zhao Huijuan;Zhang Qun;Zhu Yi;Cui Xinhong. Photosynthetic Performance and Variation in Leaf Anatomic Structure of Betula microphylla var. paludosa under Different Saline Conditions [J]. Scientia Silvae Sinicae, 2011, 47(10): 29-36. |
| [14] | Yang Fan;Ding Fei;Du Tianzhen. DREB Gene Expression in Leaves of Broussonetia papyrifera Seedlings under Salt Stress Detected by Real-Time Fluorescent Quantitative PCR [J]. Scientia Silvae Sinicae, 2010, 46(4): 146-150. |
| [15] | Ke Yuzhou;Zhou Jinxing;Zhang Xudong;Sun Qixiang;Zuo Li. Effects of Salt Stress on Photosynthetic Characteristics of Mulberry Seedlings [J]. Scientia Silvae Sinicae, 2009, 12(8): 61-66. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||